Largest Genetic Screen Resource For Cancer Established With Collation and Analysis Of CRISPR-Cas9 Datasets
Source: Thailand Medical News Dec 22, 2019 6 years, 2 months, 5 days, 4 hours, 50 minutes ago
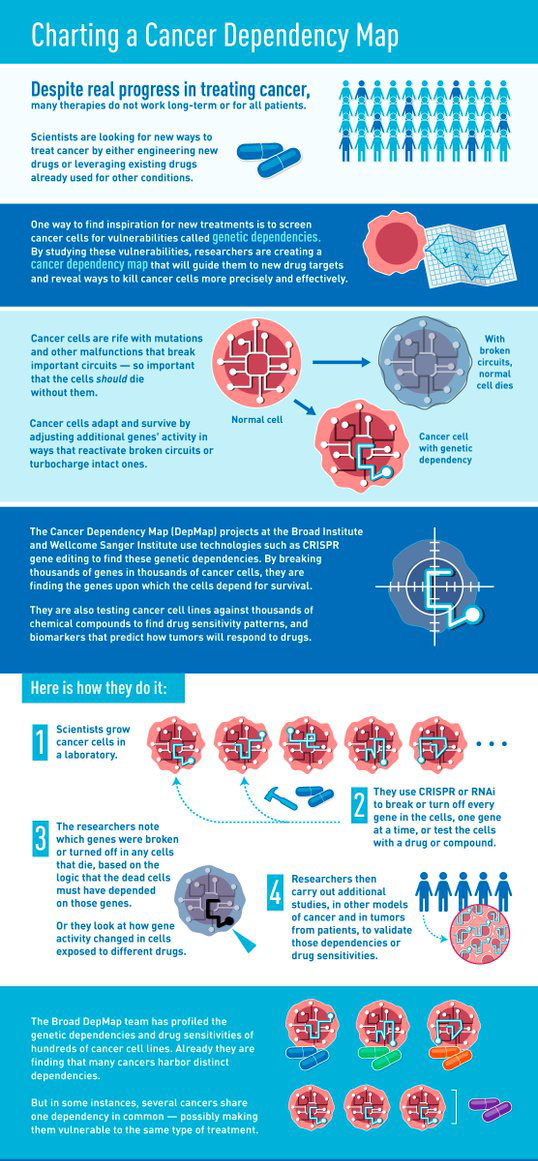
A detailed collation that can lead to a comprehensive map of genes necessary for
cancer survival is one step closer, following the validation of the two largest
CRISPR-Cas9 genetic screens in 725
cancer models, across 25 different cancer types. Scientists at the Wellcome Sanger Institute and the Broad Institute of MIT and Harvard compared the consistency of the two datasets, independently verifying the methodology and findings.

The research results, published in
Nature Communications, mean that the two datasets can be integrated to form the largest
genetic screen of
cancer cell lines to date, which will provide the basis for the
Cancer Dependency Map in around 1,000
cancer models. The scale of this combined dataset will help to speed up the discovery and development of new
cancer drugs.
The
Cancer DepMap (
Cancer Dependency Map) initiative aims to create a detailed rulebook of precision
cancer treatments for patients. Two key elements of the
Cancer DepMap are the mapping of the genes critical for the survival of cancer cells and analytics of the resulting datasets. Despite recent advances in
cancer research, making precision medicine widely available to
cancer patients requires many new drug targets.
In order to find these drug targets,
Cancer DepMap researchers take tumour cells from patients to create cell lines that can be grown in the laboratory. They then use
CRISPR-Cas9 technology to edit the genes in these
cancer cells, turning them off one-by-one to measure how critical they are for the
cancer to survive. The results of these experiments indicate which genes are the most likely to make viable drug targets.
Researchers analysed data in this new study, from two recently published
CRISPR-Cas9 genetic screens performed on
cancer cell lines at the Broad and Sanger Institutes. Despite significant differences in experimental protocols, the team found that the screen results were consistent with one another. Crucially, the same genes essential to
cancer survival known as dependencies were found in both datasets.
"The Sanger and Broad Institute
CRISPR-Cas9 screens were created using slightly different protocols, such as cell growth duration and reagents used. To verify each Institute's dataset, we have repeated
CRISPR-Cas9 screens using the protocols originally employed at the other Institute. Importantly, we have found the same genetic dependencies in each, meaning the new drug targets originally identified are consistent." commented Dr. Clare Pacini, a first author of the study from the Wellcome Sanger Institute and Open Targets during a phone interview wi
th
Thailand Medical News.
Dr Aviad Tsherniak, of the Broad Institute of MIT and Harvard, said: "This is the first analysis of its kind and is really important for the whole
cancer research community. Not only have we reproduced common and specific dependencies across the two datasets, but we have taken biomarkers of gene dependency found in one dataset and recovered them in the other. Our analysis has been unbiased, rigorous and proves the veracity of the approach and the drug targets identified."
Results comparing two large pharmacogenomic datasets employing the
cancer models used in this study in 2013 raised concerns about the reproducibility of the experiments performed. Further independently-published analyses eventually proved the two resources to be reliable and consistent, restoring confidence in the robustness of large-scale drug screens, but the episode slowed the progress of
cancer research.
This new study validates the reproducibility of
CRISPR-Cas9 functional genetic screens in order to remove any doubt about their efficacy. It sets rigorous standards for assessing these new types of dataset, facilitating the comparison and integration of large databases of
cancer dependencies.
"It is worth remembering that when these datasets were originally produced we were dealing with a new, unproven technology. This study is important because it demonstrates the validity of the experimental methods and the consistency of the data that they produce. It also means that two large
cancer dependency datasets are compatible. By joining them together, we will have access to much greater statistical power to narrow down the list of targets for the next generation of
cancer treatments." Said Dr. Francesco Iorio, of the Wellcome Sanger Institute and Open Targets.
Reference : Dempster, J.M., Pacini, C., Pantel, S. et al. Agreement between two large pan-cancer CRISPR-Cas9 gene dependency data sets. Nat Commun 10, 5817 (2019) doi:10.1038/s41467-019-13805-y 
