COVID-19 News: French Study Finds That SARS-CoV-2 Envelop Protein Has A Carboxy Terminal That Contains A PDZ Binding Motif That Is Pathogenetic
Nikhil Prasad Fact checked by:Thailand Medical News Team Dec 17, 2023 2 years, 2 months, 1 week, 17 hours, 9 minutes ago
COVID-19 News: The ongoing battle against the COVID-19 pandemic has brought researchers from around the world together in a collaborative effort to understand the intricacies of the SARS-CoV-2 virus. Among the many proteins encoded by the virus, the envelope protein (E) has emerged as a focal point of investigation due to its crucial role in the viral life cycle.
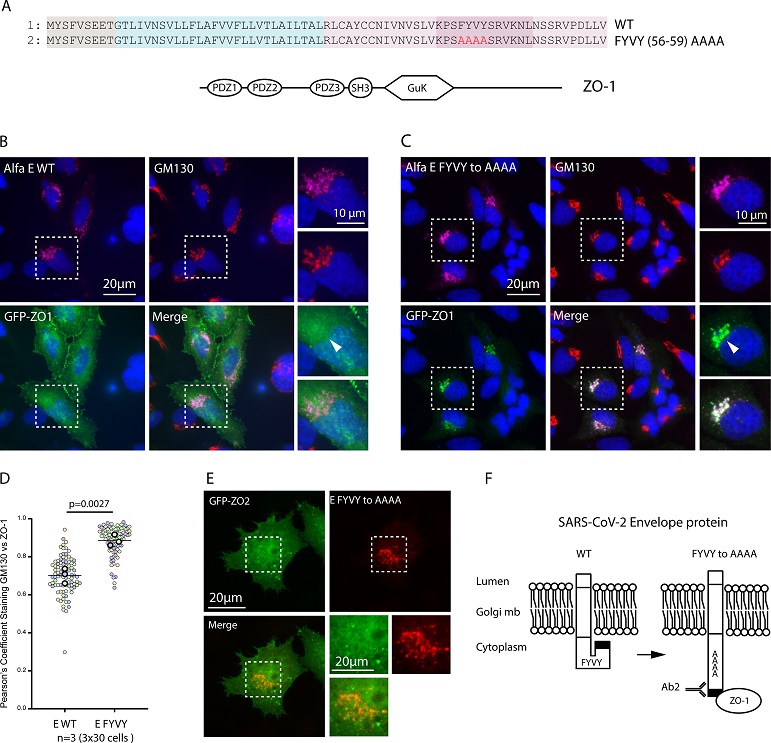 ZO-1 recruitment to E Golgi pool is regulated by the FYVY motif. (A) Sequence alignment of the WT and mutant FYVY to AAAA constructs and graphical representation of the full-length ZO-1 protein. (B) Immunofluorescence of Hela cells transfected with WT Alfa-E and full-length GFP tagged ZO-1 construct and stained with anti-Alfa and anti-GM130 antibodies. (C) Immunofluorescence of Hela Cells transfected with Alfa-E FYVY to AAAA mutant and full length GFP tagged ZO-1 construct and stained with anti-Alfa and anti-GM130 antibodies. Zoomed areas displayed are taken from the dashed square. (D) Graphs depict Pearson's Correlation coefficients computed between GFP signal and GM130 staining across three distinct biological replicates. Individual data points are represented by colors, with green, orange, and magenta corresponding to each replicate. The p value represents the result of an unpaired two-tailed t test done on the mean of the three independent experiments. (E) Immunofluorescence of Hela Cells transfected with Alfa-E FYVY to AAAA mutant and full-length GFP tagged ZO-2 construct and stained with anti-Alfa and anti-GM130 antibodies. Bar 20μm. (F) Schematic representation of the conformational shift hypothesis induced by the FYVY to AAAA mutation and its effect on Ab2 and ZO-1 binding.
ZO-1 recruitment to E Golgi pool is regulated by the FYVY motif. (A) Sequence alignment of the WT and mutant FYVY to AAAA constructs and graphical representation of the full-length ZO-1 protein. (B) Immunofluorescence of Hela cells transfected with WT Alfa-E and full-length GFP tagged ZO-1 construct and stained with anti-Alfa and anti-GM130 antibodies. (C) Immunofluorescence of Hela Cells transfected with Alfa-E FYVY to AAAA mutant and full length GFP tagged ZO-1 construct and stained with anti-Alfa and anti-GM130 antibodies. Zoomed areas displayed are taken from the dashed square. (D) Graphs depict Pearson's Correlation coefficients computed between GFP signal and GM130 staining across three distinct biological replicates. Individual data points are represented by colors, with green, orange, and magenta corresponding to each replicate. The p value represents the result of an unpaired two-tailed t test done on the mean of the three independent experiments. (E) Immunofluorescence of Hela Cells transfected with Alfa-E FYVY to AAAA mutant and full-length GFP tagged ZO-2 construct and stained with anti-Alfa and anti-GM130 antibodies. Bar 20μm. (F) Schematic representation of the conformational shift hypothesis induced by the FYVY to AAAA mutation and its effect on Ab2 and ZO-1 binding.
A recent breakthrough by researchers from the Institut Pasteur, CNRS, and Université Paris Cité-France that is covered in this
COVID-19 News report, has illuminated a previously unrecognized facet of the SARS-CoV-2 E protein - a carboxy terminal tail hosting a PDZ-binding motif (PBM), a discovery that carries profound implications for our understanding of the virus's pathogenicity.
Unraveling the Molecular Interplay
The carboxy terminal tail of the SARS-CoV-2 envelope protein has now taken center stage in the scientific spotlight, revealing a PDZ-binding motif (PBM) that plays a pivotal role in the virus's pathogenicity. PDZ-binding motifs are recognized for their ability to interact with PDZ domain-containing proteins, influencing various cellular processes. This discovery prompts a closer examination of the molecular mechanisms governing the presentation of the PBM to host PDZ domain-containing proteins, unlocking new insights into the virus-host interaction dynamics.
Conformational Regulation and Immunodetection
The Golgi apparatus, a cellular organelle, serves as a key battleground during SARS-CoV-2 infection. The study shows that at the Golgi apparatus level, the PDZ-binding motif of the E protein eludes detection by specific antibodies and the PDZ domain-containing protein binding partner. Strikingly, the introduction of four alanine substitutions upstream of the PBM induces im
munodetection by anti-E antibodies and robust recruitment of the PDZ domain-containing protein into the Golgi organelle. This finding leads researchers to propose that the presentation of the PBM to the cytoplasm is under conformational regulation, a process mediated by the central region of the E protein tail.
Viroporins and Therapeutic Implications
Viroporins, a class of small hydrophobic viral proteins capable of forming pores in host cell membranes, have emerged as key players in the viral life cycle. The SARS-CoV-2 viral genome contains three viroporin-encoding genes, including the envelope E, the ORF3a, and the ORF8. These proteins play multifunctional roles in viral replication, genome assembly, entry, and release of viral particles into infected cells. Understanding the critical role of viroporins has positioned them as attractive targets for the development of antiviral therapies.
Exploring Conservation and Interactions
The conservation of the SARS-CoV-2 E protein, particularly its high similarity between SARS-CoV-1 and SARS-CoV-2 viruses (94.7%), underscores its significance in the viral life cycle. The study reveals an association between the E protein's PDZ-binding motif and PDZ domain-containing proteins, shedding light on the intricate interplay between viral and host proteins. This interaction provides a potential avenue for viruses to exploit host cellular processes, ultimately enhancing their replication and dissemination.
Role of PDZ Domain-Containing Proteins in Viral Infection
PDZ domain-containing proteins, crucial in biological processes such as cell junction formation, cell polarity establishment, signal transduction, metabolism, and immune system signaling, are targeted by viruses to enhance their replication. The study demonstrates that infections with recombinant SARS-CoV-1 viruses lacking an E protein PBM are attenuated in mice, accompanied by a decrease in inflammatory cytokine expression and a substantial increase in survival. Conversely, mice infected with viruses containing the E protein PBM experience rapid mortality. This highlights the pivotal role of the E protein's PBM sequence in the virulence of the virus, raising questions about its specific functional role.
Immunodetection and Localization Patterns
The researchers utilized immunofluorescence experiments to elucidate the localization patterns of the SARS-CoV-2 E protein during infection. The use of novel antibodies revealed distinct Golgi and lysosomal compartmentalization, providing a nuanced understanding of the subcellular dynamics of the viral protein. Furthermore, the study explored the role of lysosomal activity in enhancing the accessibility of the E protein's carboxy-terminal extremity, adding a layer of complexity to the viral lifecycle.
Identification of Sequence Governing PBM Presentation
The exploration of the distinctive locations of the E protein led to the identification of a specific sequence, FYVY, within the viroporin that regulates the presentation of its PDZ-binding motif to the cytoplasm. Mutations in this sequence, particularly the FYVY to AAAA mutation, were found to enhance the accessibility of the E protein's C-terminal residues, facilitating immunodetection and PDZ domain binding.
Conformational Dynamics and Viral Pathogenesis
The study proposed three hypotheses to explain the regulation of PBM exposure: amino acid side chain modifications, selective cleavage of the C-terminal sequence, or an internal conformational switch. Notably, the FYVY to AAAA mutation induced a conformational change in the E protein, favoring an alpha-helix structure. The researchers postulated that this conformational switch regulates the exposure of the PBM to the cytoplasm, influencing viral pathogenesis.
Conclusion
In conclusion, the recent research conducted by French scientists has provided a comprehensive understanding of the intricate molecular mechanisms governing the SARS-CoV-2 envelope protein's PDZ-binding motif. This groundbreaking study not only sheds light on the virus's pathogenicity but also opens avenues for potential therapeutic interventions targeting viroporins and their interactions with host proteins. As the scientific community continues to unravel the complexities of COVID-19, these findings contribute significantly to our knowledge of the virus and its interactions within the host cell.
The study's implications extend beyond the laboratory, offering potential targets for antiviral therapies. The identification of the PDZ-binding motif in the E protein presents an opportunity for the development of targeted interventions that disrupt the interaction between the virus and host PDZ domain-containing proteins. By understanding the conformational dynamics and regulatory mechanisms governing the exposure of the PBM, researchers may pave the way for innovative therapeutic strategies to combat SARS-CoV-2 and other related viruses.
Moreover, the conservation of the E protein across different strains of coronaviruses highlights its evolutionary significance. The high degree of similarity between SARS-CoV-1 and SARS-CoV-2 underscores the conservation of key viral components, emphasizing the critical role of the E protein in the viral life cycle.
This knowledge can inform future research into broad-spectrum antiviral approaches that target conserved elements shared among coronaviruses.
The study's findings also raise intriguing questions about the potential role of viroporins, particularly the E protein, in the later stages of the viral life cycle. The proposal that the PBM becomes exposed to the cytoplasm at later stages, possibly during viral egress, adds a temporal dimension to our understanding of viral replication and dissemination. This insight may guide future studies investigating the intricate orchestration of events leading to the release of viral particles from infected cells.
As the global scientific community continues to collaborate and share insights, the recent study on the SARS-CoV-2 E protein adds a valuable piece to the puzzle of COVID-19. Unraveling the molecular dance between the virus and host cell components not only enhances our understanding of viral pathogenesis but also provides a foundation for the development of targeted therapeutics. The journey to conquer COVID-19 is a collective effort, and each revelation brings us one step closer to unraveling the mysteries of this formidable virus.
The study findings were published in the peer reviewed Journal of Biological Chemistry.
https://www.sciencedirect.com/science/article/pii/S0021925823026030
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
