Nikhil Prasad Fact checked by:Thailand Medical News Team Nov 07, 2023 2 years, 3 months, 4 weeks, 12 hours, 41 minutes ago
COVID-19 News: The COVID-19 pandemic, caused by the SARS-CoV-2 virus, has gripped the world for over two years, leading to unprecedented challenges for public health. The search for answers and solutions to this global crisis has been relentless, and scientists have explored various aspects of the virus, including its genetic basis. One crucial aspect of this exploration is understanding the role of host genetics in COVID-19 outcomes. This
COVID-19 News article delves into a groundbreaking study conducted by researchers at Shanghai Jiao Tong University School of Medicine-China, the First People's Hospital of Wenling, Taizhou-China, and the Chinese Academy of Sciences, Shanghai-China. Their research integrates data from genome-wide association studies (GWAS), host genetics, and RNA interactomes to identify two risk genes, FUBP1 and RAB2A, as key factors in the development and progression of COVID-19.
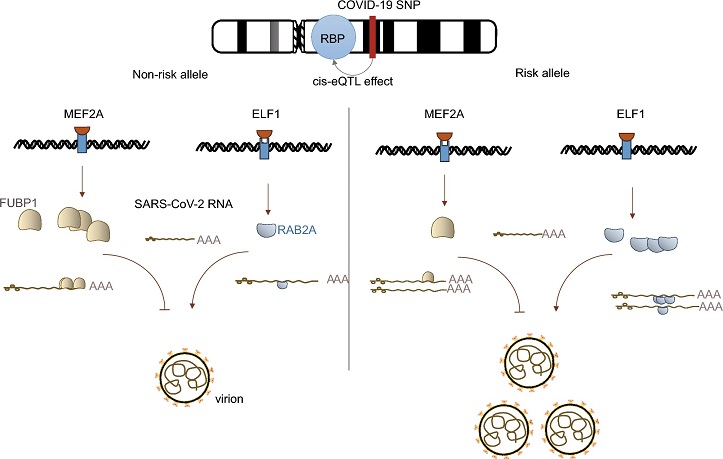 Hypothesis model of risk variants mapped to RBPs predispose hospitalization of COVID-19. MEF2A strongly binds to the non-risk allele of FUBP1 and ELF1 weakly binds to that of RAB2A, which enhances the anti-viral ability of FUBP1 and reduces the pro-viral capacity of RAB2A, leading to less virion. In contrast, MEF2A weakly binds to the risk allele of FUBP1, and ELF1 strongly binds to that of RAB2A, which reduces the anti-viral ability of FUBP1 and enhances the pro-viral capacity of RAB2A, leading to more virion and worse outcome of COVID-19.
Genome-Wide Association Studies (GWAS)
Hypothesis model of risk variants mapped to RBPs predispose hospitalization of COVID-19. MEF2A strongly binds to the non-risk allele of FUBP1 and ELF1 weakly binds to that of RAB2A, which enhances the anti-viral ability of FUBP1 and reduces the pro-viral capacity of RAB2A, leading to less virion. In contrast, MEF2A weakly binds to the risk allele of FUBP1, and ELF1 strongly binds to that of RAB2A, which reduces the anti-viral ability of FUBP1 and enhances the pro-viral capacity of RAB2A, leading to more virion and worse outcome of COVID-19.
Genome-Wide Association Studies (GWAS)
The foundation of this study lies in the field of genome-wide association studies (GWAS), a powerful tool for unraveling the genetic underpinnings of complex traits and diseases. Numerous genetic variants associated with COVID-19 have already been identified using GWAS, including the notable variant 12q24.13 located in OAS1, which influences COVID-19 susceptibility and severity. While GWAS has made significant progress in uncovering these genetic links, challenges persist, particularly in translating these findings into clinical applications, especially in cases where the implicated variants are located in non-coding regions. Additionally, understanding how these genetic variants impact COVID-19 risk remains an ongoing challenge.
Cis-Expression Quantitative Trait Loci (cis-eQTL) Analysis
Cis-expression quantitative trait loci (cis-eQTL) analysis provides a valuable tool to bridge the gap between GWAS findings and gene expression. By associating genetic variants with gene expression, cis-eQTL analysis helps identify risk variants for COVID-19 that influence the expression levels of specific genes. For instance, Horowitz et al. discovered a genetic variant, rs190509934, which protects against SARS-CoV-2 infection and downregulates the expression of ACE2, a gene crucial for the virus's entry into host cells. This approach sheds light on the impact of non-coding variants on gene expression levels, enhancing our understanding of the genetic risk factors associated with COVID-19.
The Complex Interplay Between SARS-CoV-2 and Host Genetics
SARS-CoV-2 is a highly adaptable virus that relies on intricate interactions with host cellular machinery to
complete its viral life cycle and evade the host's defense mechanisms. These interactions occur at various levels, from protein synthesis to the RNA-dependent RNA polymerase (RdRp) complex and the manipulation of host cellular pathways to promote viral replication. On the flip side, the host cells activate anti-viral detection and defense mechanisms in response to SARS-CoV-2 infection, employing proteins like RIG-I, MDA5, and ZAP to recognize and combat the virus.
RNA Binding Proteins (RBPs) in the SARS-CoV-2 RNA Interactome
Among the key host cellular factors interacting with RNA viruses are RNA binding proteins (RBPs). These versatile proteins play a pivotal role in various aspects of biological processing, including viral replication, RNA metabolism, RNA stability, and translation. Host RBPs that interact with SARS-CoV-2 RNA have been identified as key players in the virus's life cycle. For example, CNBP and LARP1 restrict SARS-CoV-2 replication, while IGF2BP1 stabilizes and enhances the translation of SARS-CoV-2 RNA.
https://www.thailandmedical.news/news/breaking-scientists-warns-that-sars-cov-2-also-interacts-with-igf2bp1-an-oncofetal-rna-binding-protein-that-typically-fuels-tumor-virus-propagation
However, previous studies on RBPs interacting with SARS-CoV-2 RNA were primarily conducted on cell lines, limiting their ability to accurately replicate human infections and establish a comprehensive SARS-CoV-2 RNA interactome. Therefore, it is essential to link host genetic risk variants for COVID-19 with the expression levels of RBPs that interact with SARS-CoV-2 RNA and explore the mechanisms underlying these interactions, providing substantial evidence for the role of RBPs in COVID-19.
Identification of FUBP1 and RAB2A as Risk Genes
In their groundbreaking study, the researchers focused on the SARS-CoV-2 RNA interactome within the lung, utilizing data from the COVID-19 Host Genetics Initiative (HGI). This initiative involved annotating all genetic variants, including those in high linkage disequilibrium (LD), and categorizing them as either coding or non-coding variants. To identify risk genes associated with COVID-19, the study incorporated genes with nonsynonymous mutations and expression quantitative trait loci (eGENEs) within the lung tissue.
The researchers further investigated the interaction between host RBPs and SARS-CoV-2 RNA, which plays a critical role in various stages of the virus's life cycle. This led to the identification of three RBP-related loci associated with COVID-19 risk. To validate the significance of these findings, the researchers analyzed single-cell RNA sequencing data, revealing the expression of FUBP1 and RAB2A in SARS-CoV-2-infected upper respiratory tract epithelial cells.
Functional Variants and Their Impact
The study also focused on specific genetic variants and their functional significance. By surveying allele-specific transcription factor sites and cis-regulatory elements and conducting motif analysis, the researchers identified two functional variants: NC_000001.11:g.77984833C>A and NC_000008.11:g.60559280T>C. These variants had a profound impact on the transcription factor binding affinity, leading to the downregulation of FUBP1 and the upregulation of RAB2A. Consequently, patients with these risk alleles exhibited altered expression levels of these genes, shedding light on the mechanisms by which these genetic variants influence COVID-19 outcomes.
The Role of FUBP1 and RAB2A in COVID-19
FUBP1, a canonical RBP, belongs to a family of single-stranded DNA-binding regulators known as FUBPs. While primarily located in the nucleus, there is evidence to suggest that FUBP1 may also be expressed in the cytoplasm, where it regulates cytoplasmic virus RNA. Studies have shown a significant downregulation of FUBP1 in COVID-19 patients, particularly in peripheral blood monocytes. Though the exact function of FUBP1 in COVID-19 is not fully understood, previous research has indicated that FUBP1 can suppress protein translation of other viruses, such as the Japanese encephalitis virus (JEV), by targeting its 5' and 3' untranslated regions (UTRs). Given that FUBP1 binds to SARS-CoV-2, it is hypothesized that FUBP1 exerts an anti-viral effect by suppressing viral transcription or protein translation, possibly through its interaction with specific regions of the SARS-CoV-2 RNA. Therefore, targeting FUBP1 could hold promise in the treatment of COVID-19, with research suggesting that FUBP1 expression could be restored after exposure to certain compounds, such as allicin, in SARS-CoV-2-infected cells.
RAB2A, on the other hand, is a member of the Rab family and was identified as an RBP relatively recently. Although its precise role in virus RNA is not fully elucidated, RAB2A is well-established as a critical modulator of intracellular membrane trafficking, particularly in protein transport within the ER-Golgi intermediate compartment (ERGIC). This process is crucial for the assembly and transport of structural and non-structural viral proteins. Recent research has shown that RAB2A interacts with several components of SARS-CoV-2, including NSP7 and ORF3a, which could indicate its role in viral replication and evasion of the host's immune response. Moreover, RAB2A has been identified as a SARS-CoV-2 binding protein, suggesting that it might contribute to the stabilization of SARS-CoV-2 RNA by forming a complex that binds to both viral RNA and NSP7. This interaction could facilitate viral replication, making RAB2A a potential pro-viral factor in COVID-19. The meta-analysis of GWAS and Mendelian randomization analyses have also supported the role of RAB2A in severe COVID-19. Searching through the ChEMBL database, the researchers identified CID1067700 as a potential inhibitor of RAB2A, offering a potential avenue for targeting this RBP in the treatment of COVID-19.
Potential Therapeutic Approaches
While few drugs specifically target FUBP1 and RAB2A, the research suggests that using RNA-binding proteins as therapeutic targets for COVID-19 is promising. For example, pioglitazone and lapatinib have been identified as potential drugs that influence the expression of specific RBPs. Another study has investigated drugs targeting proteins that interact with SARS-CoV-2-related RBPs and identified potential candidates, such as Doxorubicin and Topotecan. These drugs offer alternative strategies for mitigating the impact of COVID-19 by targeting the protein interactome of RBPs.
Limitations and Future Directions
Despite these significant findings, some limitations and avenues for future research should be acknowledged. The exact binding sites of RAB2A on SARS-CoV-2 RNA were not identified due to limited CLIP-seq data. Further investigations using RNA Binding Protein Immunoprecipitation Assay Sequencing (RIP-seq) and Gel Shift Assays (EMSA) are warranted to pinpoint these binding sites and analyze the effects of genetic variations on SARS-CoV-2 RNA binding. Moreover, the study did not delve into the detailed mechanisms by which specific variants, such as NC_000001.11:g.77984833C>A and NC_000008.11:g.60559280T>C, regulate the expression levels of FUBP1 and RAB2A. Future experiments, including CRISPR-mediated inhibition (CRISPRi) and activation (CRISPRa), could help validate the genetic effects of these variants on RBPs, and ChIP-Seq (or ChIP-qPCR) could investigate the binding affinity of transcription factors. The study also leaves room for further exploration of the precise roles played by FUBP1 and RAB2A in SARS-CoV-2 infection.
Conclusion
In summary, this groundbreaking study seamlessly integrates host genetics, RNA interactomes, and GWAS data to unravel the genetic mysteries of COVID-19. By linking genetic variants to the expression of RNA binding proteins that interact with SARS-CoV-2 RNA, the research identifies FUBP1 and RAB2A as crucial risk genes for the development and severity of COVID-19. These findings offer valuable insights into the genetic risk factors underpinning COVID-19, shedding light on the roles of FUBP1 and RAB2A as RNA binding proteins in the context of SARS-CoV-2 infection. The study also proposes a model suggesting that genetic variants may influence the binding affinity of transcription factors, thereby altering the expression levels of RBPs and impacting their function in the virus's life cycle. These findings open new avenues for understanding the complex interplay between genetics and COVID-19 and offer a potential roadmap for the development of targeted therapeutic approaches in the fight against this global pandemic.
The study findings were published in the peer reviewed journal: Scientific Reports.
https://www.nature.com/articles/s41598-023-44705-3
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
