COVID-19-Genetics: Regulatory Variants Of APOBEC3 Genes Linked To COVID-19 Severity In Populations With African Ancestry
Thailand Medical News Team Aug 19, 2023 2 years, 5 months, 3 weeks, 5 days, 4 hours, 26 minutes ago
COVID-19-Genetics: In a groundbreaking study poised to reshape our understanding of the COVID-19 pandemic, researchers from Guizhou Medical University-China, in collaboration with The Affiliated Hospital of Guizhou Medical University-China and St. Jude Children’s Research Hospital-USA, have shed light on a potential genetic connection between regulatory variants of APOBEC3 genes and the severity of COVID-19 in populations with African ancestry.
 Pic Credit: Shutterstock
Pic Credit: Shutterstock
The
COVID-19-Genetics study, which delves deep into the intricate genetic landscape of the virus, could pave the way for more effective strategies in controlling the ongoing pandemic.
Since its ominous emergence in November 2019, the SARS-CoV-2 virus has wreaked havoc on global populations, plunging nations into a relentless battle against a formidable adversary. As the virus continues its unrelenting assault, researchers have grappled with the ever-evolving mutations within the SARS-CoV-2 genome. These mutations, often characterized by the nucleotide substitution C > U, have posed significant challenges in efforts to control and combat the virus's spread.
A particular spotlight has fallen on the human host factors APOBEC3 genes, known for their involvement in catalyzing the editing of viral genetic material. These genes have been implicated in the enrichment of C > U mutations in SARS-CoV-2 sequences, raising intriguing questions about their potential influence on the virus's evolution.
The study takes a remarkable turn by examining the variants Beta (B.1.351) and Omicron (B.1.1.529), which originated in South Africa and sent ripples of concern across the globe.
The study team postulated that unique genetic variants of APOBEC3 genes in African populations might be instrumental in driving the higher mutation rate observed in SARS-CoV-2 variants emanating from the African continent. This hypothesis laid the foundation for an in-depth investigation into the association between APOBEC3 genes and COVID-19 severity in African populations.
To unveil the intricate genetic interactions at play, the study team embarked on a comprehensive analysis, drawing from an array of data sources, including the 1000 Genomes Project, GTEx, and the Host Genetics Initiative of COVID-19. The integration of these diverse datasets enabled the identification of potential functional single-nucleotide polymorphisms (SNPs) situated proximal to APOBEC3 genes. Notably, these SNPs exhibited a robust association with COVID-19 hospitalization exclusively within African populations.
Among the triumvirate of SNPs singled out by the study, rs12168809 and rs76929059, nestled in the intergenic and promoter region of APOBEC3A, stood out as polymorphic entities exclusive to African populations.
These SNPs also acted as expression quantitative trait loci (eQTLs) for multiple APOBEC3 genes across various tissues. The third SNP, rs13057307, emerged as a risk allele for COVID-19 hospitalization in s
amples with European ancestry, showcasing differential minor allele frequencies across African populations.
This revelation presents a compelling narrative about the pivotal role of APOBEC3 genes in the intricate dance between the virus and its human host. These genes, well-known for their involvement in combating viral infections, have emerged as key players in the hypermutation of SARS-CoV-2. By deaminating specific bases in the viral RNA, APOBEC3 genes wield a potent influence on the virus's genetic makeup, ultimately shaping its evolutionary trajectory.
The study further illuminates the complex nature of APOBEC3 gene family members, each wielding unique properties that contribute to the host's defense against viral invaders. APOBEC3B, expressed predominantly in the nucleus, emerges as a potent modulator of host immunity, influencing viral replication and aiding in the suppression of vesicular stomatitis virus. Conversely, APOBEC3D, situated in the cytoplasm, exerts its influence on HIV-1 genome diversification, contributing to the virus's remarkable adaptability.
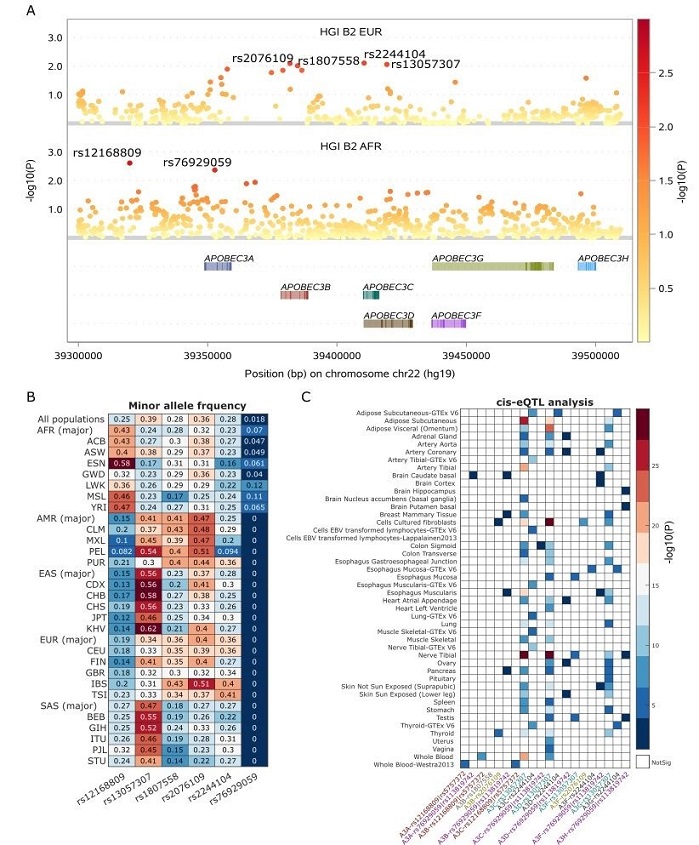 Association of single nucleotide polymorphisms (SNPs) of APOBEC3 genes with COVID-19 hospitalization. (A) Local Manhattan plots demonstrate that 6 SNPs close to APOBEC3 genes display nominally association signals (P< 0.01) in two COVID-19 hospitalization GWASs with different ancestries, including HGI-B2 of European American and African American. Among these 6 COVID-19 risk SNPs, 2 SNPs are specific derived from COVID-19 hospitalization GWAS with African ancestry and the other 4 SNPs are unique to COVID-19 hospitalization GWAS with European ancestry. (B) Population frequencies of these 6 SNPs across 5 major populations, including African (AFR), Admixed American (AMR), East Asian (EAS), South Asian (SAS), and European (EUR), as well as among all or sub-populations of these major populations are illustrated in a heatmap. Each heatmap cell displays the population frequency of each corresponding SNP. The AFR-specific COVID-19 risk SNP rs76929059 is only polymorphic in AFR populations. (C) cis-eQTL analysis for these 6 COVID-19 risk SNPs or its high linkage disequilibrium (R2 > 0.9 in EUR or AFR populations) across multiple tissues in GTEx and other eQTL databases.
Association of single nucleotide polymorphisms (SNPs) of APOBEC3 genes with COVID-19 hospitalization. (A) Local Manhattan plots demonstrate that 6 SNPs close to APOBEC3 genes display nominally association signals (P< 0.01) in two COVID-19 hospitalization GWASs with different ancestries, including HGI-B2 of European American and African American. Among these 6 COVID-19 risk SNPs, 2 SNPs are specific derived from COVID-19 hospitalization GWAS with African ancestry and the other 4 SNPs are unique to COVID-19 hospitalization GWAS with European ancestry. (B) Population frequencies of these 6 SNPs across 5 major populations, including African (AFR), Admixed American (AMR), East Asian (EAS), South Asian (SAS), and European (EUR), as well as among all or sub-populations of these major populations are illustrated in a heatmap. Each heatmap cell displays the population frequency of each corresponding SNP. The AFR-specific COVID-19 risk SNP rs76929059 is only polymorphic in AFR populations. (C) cis-eQTL analysis for these 6 COVID-19 risk SNPs or its high linkage disequilibrium (R2 > 0.9 in EUR or AFR populations) across multiple tissues in GTEx and other eQTL databases.
Crucially, the study underlines the urgent need for further investigation and replication to validate the observed associations. While the identified SNPs offer tantalizing insights into the potential impact of APOBEC3 genes on COVID-19 severity, direct experimentation and replication efforts are imperative to unravel the complex web of interactions and confirm the exact role of APOBEC3 genes in the generation of high-transmissibility SARS-CoV-2 variants.
As the world grapples with the persistent challenges posed by the COVID-19 pandemic, this pioneering study offers a beacon of hope. By unravelling the intricate interplay between host genetics and viral evolution, researchers inch closer to deciphering the enigma of SARS-CoV-2 rapid evolution and the generation of many mutations. The potential implications of these findings are far-reaching, offering new avenues for targeted interventions, treatment strategies, and a deeper comprehension of the virus that has held the world captive for years.
The study findings were published on a preprint server and are currently being peer reviewed for publication into the journal: Scientific Reports.
https://www.researchsquare.com/article/rs-3171718/v1
For the latest on
COVID-19-Genetics, keep on logging to Thailand Medical News

