Study Focusing On The Mutations Of The S Glycoproteins Of The Omicron Variants Reveals That It Is More Infectious And Immune Evasive!
Source: Omicron Variant Dec 07, 2021 4 years, 2 months, 2 weeks, 3 days, 13 hours, 24 minutes ago
Omicron Variant: Yet another study, this time by researchers from the ICMR-National Institute of Cholera and Enteric Diseases, Kolkata-India focusing on the mutations of the S glycoproteins of the new Omicron variant has revealed that the new variant is indeed more infectious and immune evasive.
The study team performed a comprehensive analysis of the S glycoprotein mutations of 309 strains of the Omicron variant and also discussed the probable effects of observed mutations on several aspects of virus biology based on known available knowledge of mutational effects on S glycoprotein structure, function, and immune evasion characteristics.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.medrxiv.org/content/10.1101/2021.12.04.21267284v2
As the SARS-CoV-2 coronavirus continues to circulate around the world, multiple new mutations continue to emerge. These have led to the detection of new variants, including the Alpha, Beta, and Delta variants that have been shown to be more transmissible, more capable of immune evasion, or both, compared to the ancestral variants.
To make worse, the Delta variant is now spawning numerous sub-variants, each even their own sub-lineages and to date, almost 200 odd delta-sub-variants have been identified, each with unique and concerning mutations.
Near the end of November 2021, the latest new variant called B.1.1.529 or the
Omicron variant made its debut and was discovered in South Africa as a result of genome sequencing surveillance. With over 30 new mutations and markedly higher transmissibility, scientists are wondering if this could drive devastating new surges of reinfection, overcoming the immunity elicited by earlier infections or by current vaccines.
This new study explores the spike glycoprotein mutations in the Omicron variant, with their putative effects on the biological characteristics of the virus.
The B.1.1.529 or Omicron variant, was first designated a variant of concern (VOC) by the World Health Organization (WHO) based on the presence of numerous mutations, some of which are common to earlier VOCs. This meant that the Omicron variant was likely to be more infectious, cause more severe disease, escape neutralization by antibodies elicited by earlier variants or vaccines, and be more difficult to diagnose and/or treat.
Alarmingly the detection of the Omicron variant in South Africa occurred against a backdrop of rapidly rising cases, but further genomic sequencing studies are required to tell if this variant is driving the newest surge in infections. The earliest evidence of this variant is in a sample collected on November 9th, 2021.
It should be noted that the viral spike is the surface protein that mediates viral attachment to the host cell angiotensin-converting enzyme 2 (ACE2) receptor, thereby making viral entry into the host cell possible. It is therefore also the prime target of neutralizing antibodies to the virus.
The SARS-CoV-2 spike glycoprotein has two subunits, the S1 and S2, which mediate ACE2 attachment and membrane fusion, respectively, to a
ccomplish viral entry into the target host cell. These two subunits require to be cleaved by a host furin enzyme and then show non-covalent interactions.
The receptor-binding domain or RBD of the S1 subunit is involved in ACE2 receptor engagement, while the N-terminal domain or NTD recognizes several attachment factors. The S2 subunit contains the viral-host cell membrane fusion apparatus, which shows a large conformational change to cause fusion to occur. This allows the viral genome to enter the target cell via endocytosis, paving the way for productive infection.
Importantly the RBD is immunodominant, eliciting the majority of protective and binding antibodies to the virus. Since it also accomplishes virus-host cell receptor engagement, an RBD mutation is especially likely to affect the ability of antibodies to neutralize the spike. Such antibodies, elicited by vaccination or as the result of natural infection, are the basis of immune protection against infection.
Furthermore, RBD mutations could also, conceivably, impact virus-receptor binding. These facts prompted the current study to identify the mutations and predict their effect on viral transmissibility, pathogenicity, and immune evasion. This prediction is necessarily based on what is known about the earlier identified mutations.
Typically, as the virus circulates in a large pool of susceptible individuals, some with antibodies against earlier strains as a result of prior infection or vaccination, mutations that change the shape or behavior of the spike in a manner that benefits the virus tend to be propagated.
For example, if the mutation facilitates immune evasion or immune escape or allows more rapid infection to take place, it would be more likely to persist and become part of a new variant that may become dominant.
For this reason, spike mutations have been the focus of all classification and functional studies of the variants of the virus.
The present study not only analyses the mutations in the Omicron spike variant but classifies various combinations of spike mutations into groups.
The Global Initiative for Sharing Avian Influenza Data (GISAID) database now contains over 300 Omicron sequences from African, European, and Asian countries, mostly from South Africa and other nearby countries.
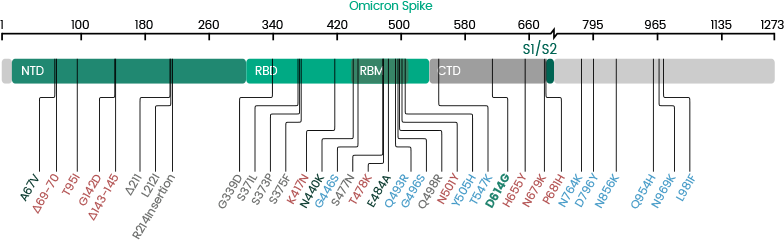
The study team mapped the 37 spike mutations of the Omicron variant, which are present in anywhere between 60% to 100% of the sequences available so far. Of these, 29 occur in the S1 domain, comprising 11 in the N-terminal domain (NTD), 15 in the receptor-binding domain (RBD), and 3 in the C-terminal domain (CTD). Of the 15 RBD mutations, ten will be on the receptor-binding motif (RBM). In contrast, eight mutations were on the S2 domain.
Interestingly, of the 37 mutations, 25 were found to be unique to this strain, while 12 are shared with the Alpha, Beta, Gamma, and Delta VOCs.
All the 300-plus strains of the Omicron were classified into 60 groups, each with its own set of mutations. Group 1 and group 2 contain more than half the total number, at 170, containing all 37 mutations and 33 mutations, respectively. Group 1 contains almost 120 of these sequences, and Group 2 about 50.
The remaining approximate 140 sequences were classified into the remaining 60 groups.
 Importantly the presence of so many groups shows the multiplicity of mutational subtypes of this variant.
Importantly the presence of so many groups shows the multiplicity of mutational subtypes of this variant.
It should be noted that phylogenetically, a new cluster of the Omicron variant, comprising 155 strains, emerged from the GR, Pango B.1.1 or 20B clade of SARS-CoV-2. This cluster is further subdivided into smaller clusters by spike mutation type.
Alarmingly the presence of 37 mutations in the spike protein, especially affecting the crucial NTD and RBD portions of the S1 subunit of the protein, has raised concerns about a new wave of infections.
The 12 overlapping mutations have been shown to cause higher transmissibility, higher binding affinity, or immune evasion.
Numerous scientists have predicted that the Omicron variant will continue to show these characteristics.
The other 25 unique mutations are still unknown concerning their biological impacts.
Importantly the 4 RBD mutations K417N, K477N, T478K, and E484A, have been shown to confer immune evasion capabilities.
It was found that both T478K and E484A affect residues at the immunodominant site of the RBD. The E484K mutation emerges in patients who receive monoclonal antibody or convalescent plasma therapy and confers immune escape capability. Other mutations at this site, such as E484A, E484D, and E484G, also resist antibody-mediated neutralization, while E484Q reduces the titer of serum neutralizing antibodies.
Worryingly the K477N mutation enables the variant to resist neutralization by monoclonal antibodies but not convalescent plasma. The K417N, previously identified in the Beta and Delta plus variants, makes them partially resistant to neutralization by therapeutic monoclonal antibodies raised against earlier variants.
Significantly, the fact that these three immune escape mutations are found in the Omicron variant may lead to a heightened potential for immune escape.
The N-terminal domain or NTD is also immunogenic and contains an antigenic supersite comprising the N-terminal end, a β-hairpin, and loop residues. The Omicron variant contains four mutations within the β-hairpin region, which may enhance immune evasion by this variant.
While four of the 11 mutations in the Omicron RBD, namely, G339D, N440K, T478K, and N501Y, were shown in a recent study to increase ACE2 binding affinity, and the others reduced it.
The critical among these are Q493, Q498, and N501, because they form contact networks including hotspot residues (K31 and K353) on the ACE2 contact surface. If these are replaced by nonpolar amino acids, RBD-ACE2 binding affinity increases. Conversely, replacing glutamine at positions 493 and 498 by the more polar residue arginine may reduce binding affinity.
Hence, the magnitude of interactions between these 11 and 4 mutations with opposing effects will determine the ultimate effect on the receptor binding affinity of the Omicron variant.
The significant presence of D614G and P681H mutations that mediated high transmissibility in earlier variants means that the Omicron is expected to be highly infectious.
Currently the Delta continues to spread rapidly in susceptible communities and regions, while significant levels of vaccine-induced and natural immunity also exist. Even so, the Omicron variant is causing rapid rises in the number of infections. This seems to suggest that this strain is both highly transmissible and able to cause breakthrough infections.
It is predicted that the omicron will supplant delta as the most common variation in South Africa and other part of the world very rapidly and may lead to another wave of COVID-19.
But what is also worrisome is that the Omicron is also still evolving and mutating, meaning it could get far worse considering that the manner of the mutations are not causing the virus to be weak or milder but rather in the opposite direction ie more potent, virulent and transmissible.
Please help to sustain this site and also all our research and community initiatives by making a donation. Your help means a lot and helps saves lives directly and indirectly and we desperately also need financial help now.
https://www.thailandmedical.news/p/sponsorship
For the latest on the
Omicron Variant, keep on logging to Thailand Medical News.

