COVID-19 News: Oligomer Formation Of SARS-CoV-2 ORF8 Through 73YIDI76 Motifs Regulates Immune Response And Non-Infusion Antiviral Interactions
Nikhil Prasad Fact checked by:Thailand Medical News Team Nov 29, 2023 2 years, 3 months, 1 week, 4 days, 18 hours, 19 minutes ago
COVID-19 News: The relentless pursuit of effective COVID-19 therapeutics has led researchers to explore new avenues beyond the conventional targets of spike and RNA-dependent RNA polymerase proteins. In a groundbreaking study conducted by the Faculty of Life Sciences and Biotechnology at Shahid Beheshti University in Iran, a spotlight has been cast on Open Reading Frame 8 (ORF8), a specific accessory protein of SARS-CoV-2. This 121-amino acid protein, while often overlooked, plays pivotal roles in viral infectivity and pathogenesis. This
COVID-19 News report delves into the intricate details of the oligomer formation of ORF8 through the 73YIDI76 motifs, shedding light on its regulation of immune responses and potential for innovative non-infusional antiviral interactions.
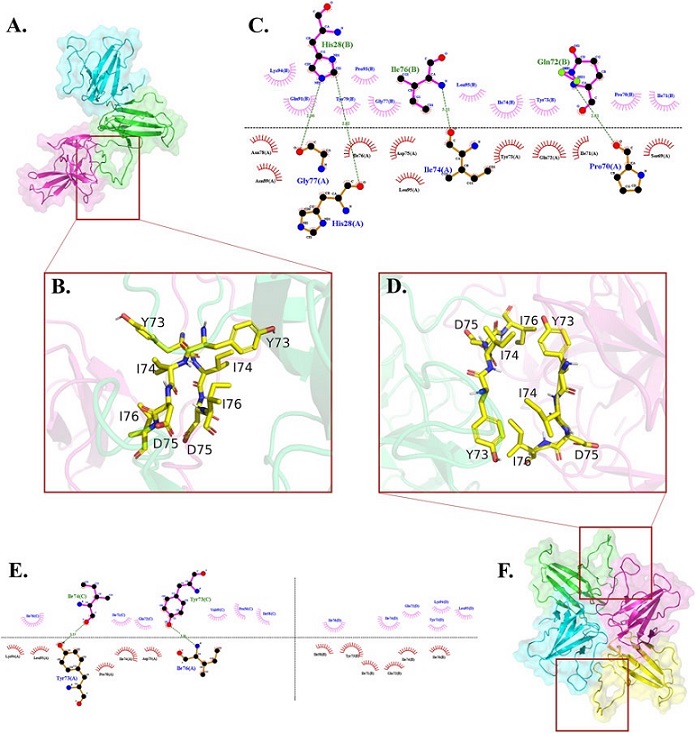 Where the 73YIDI76 motifs could be assembled differently among trimers and tetramers, the interface residues interact hydrophobically with each other. (A) Top-ranked trimer structure. (B) The 73YIDI76 motifs are shown as sticks are parallel to one another in the trimers. (C) The atomistic interactions of the trimer interface residues. (D) In the most stable tetramers, the 73YIDI76 motifs are positioned in antiparallel strands next to one another. In this illustration, one of the two dimer linking sites is shown. (E) The diagram illustrates the atomistic interactions within the non-covalent interfaces of the tetramer. (F) Top-ranked tetramer structure.
SARS-CoV-2 and the Significance of ORF8
Where the 73YIDI76 motifs could be assembled differently among trimers and tetramers, the interface residues interact hydrophobically with each other. (A) Top-ranked trimer structure. (B) The 73YIDI76 motifs are shown as sticks are parallel to one another in the trimers. (C) The atomistic interactions of the trimer interface residues. (D) In the most stable tetramers, the 73YIDI76 motifs are positioned in antiparallel strands next to one another. In this illustration, one of the two dimer linking sites is shown. (E) The diagram illustrates the atomistic interactions within the non-covalent interfaces of the tetramer. (F) Top-ranked tetramer structure.
SARS-CoV-2 and the Significance of ORF8
As of July 31, 2023, the COVID-19 pandemic has claimed over 6.9 million lives and infected 768.5 million individuals worldwide. The causative agent, SARS-CoV-2, belongs to the Betacoronavirus genus, along with its predecessors SARS-CoV and MERS. While the focus of current treatments has primarily centered on spike and RNA-dependent RNA polymerase proteins, the role of accessory proteins, especially ORF8, in the virus-host interaction network is gaining prominence.
The SARS-CoV-2 RNA genome comprises 12 open reading frames (ORFs), encoding four structural proteins, 16 nonstructural proteins (Nsps), and 11 accessory proteins. Structural proteins are vital for virion assembly, Nsps are involved in transcription and viral replication, and accessory proteins, including ORF8, have the ability to modulate the host cell's molecular network, influencing the immune system.
ORF8's Influence on Immune Responses
Research has shown that ORF8 possesses a unique correlation with the pathogenesis of SARS-CoV-2. A 382-nucleotide deletion in the ORF8 gene has been associated with milder infections in patients. Experimental studies have identified 47 human proteins as ORF8 interacting partners, making it statistically significant in the virus-host protein network, alongside the membrane (M) protein and Nsp7.
Notably, ORF8 mimics various immune molecules, such as Interleukin-17, leading to the activation of IL-17 receptors A and C and triggering a robust inflammatory response. Furthermore, ORF8 is implicated in disrupting the interferon type I signaling pathway and directly interacting with major histocompatibility complex class &Iot
a; (MHC-I), downregulating its expression. These multifaceted interactions position ORF8 as a key player in immune evasion and inflammation.
Structural Insights into ORF8 Oligomerization
The structural characteristics of ORF8, including an Ig-like domain and a flexible β4-β5 loop containing the 73YIDI76 motif, make it a dynamic player in the virus-host interaction network. Experimental observations reveal a covalently bonded dimeric structure of ORF8, maintained by intramolecular disulfide bonds. However, recent studies indicate a novel dimension to ORF8's structural dynamics - the ability to form functional multimeric assemblies, particularly tetramers, a phenomenon not observed in SARS-CoV.
The Oligomerization Mechanism
The oligomerization of ORF8 has been attributed to crystallographic contacts centered by 73YIDI76 motifs. Molecular modeling and molecular dynamics simulations were employed to model trimeric and tetrameric ORF8 oligomeric forms. Results unveiled that β4-β5 loops, specifically the 73YIDI76 motifs, play a crucial role in stabilizing trimeric and tetrameric forms. The tetramers, resembling a doughnut-like construction, emerged as the most stabilized oligomeric structures, with four β4-β5 loops forming interfaces between two dimers.
Notably, each monomer in the tetramer links to two others through β4-β5 loops and a covalent Cys20-Cys20 bridge, creating a highly stable configuration. Epitope mapping, binding site predictions, and solvent-accessible surface area analyses demonstrated that B-cell, MHC-I, and drug epitopes remain exposed in oligomeric forms, emphasizing their potential as therapeutic targets.
Dynamics and Stability of Oligomeric Forms
Molecular dynamics simulations were conducted to study the dynamics and stability of ORF8 oligomeric forms. Results indicated that doughnut-like tetramers exhibited noticeably higher stability compared to trimeric forms. The analyses, including Root Mean Square Deviation (RMSD) values and Principal Component Analysis (PCA), supported the experimental findings, highlighting the distinct stability of tetrameric structures.
Comparisons of average RMSD values between trimeric and tetrameric systems and native ORF8 dimer further elucidated the varying degrees of stability among these systems. The β4-β5 loop region and the MHC-I binding site exhibited higher Root Mean Square Fluctuation (RMSF) values, emphasizing their inherent flexibility, which plays a critical role in interactions.
Oligomeric Structure and Therapeutic Implications
The oligomeric structure of ORF8 presents a novel framework for understanding its protein-protein interactions. The 73YIDI76 motifs, exposed at the surface of the dimer, showcase high flexibility, indicating the potential for diverse conformations and interactions with various protein or ligand partners.
Interestingly, the oligomer formation has no discernible effect on the MHC-I epitopes, which remain functional. This opens up possibilities for targeted drug design, especially in disrupting oligomerization. The covalent interface and non-covalent interfaces, represented by the β4-β5 loop, emerge as potential drug target hotspots. Disrupting these interfaces could hinder oligomerization and impact the protein's pathogenesis activities.
Future Directions and Conclusion
The research from Shahid Beheshti University in Iran marks a significant leap in understanding the complex dynamics of SARS-CoV-2 ORF8. As the world grapples with the ongoing COVID-19 pandemic, the insights gained from this study pave the way for future research initiatives. Exploring the influence of inhibitors, mutations, and glycosylation on ORF8 oligomerization, as well as examining different physiological situations, holds promise for advancing our knowledge of ORF8's role in the virus-host interaction network.
In conclusion, ORF8 emerges as a critical player in SARS-CoV-2 pathogenesis, with its unique ability to form stable multimeric assemblies. The structural insights into ORF8 oligomerization provide a foundation for the development of targeted antiviral medications, offering a potential paradigm shift in COVID-19 therapeutics. As researchers delve deeper into the complexities of ORF8 and its oligomeric forms, the hope is that this knowledge will contribute to the development of more efficient and precise treatments for COVID-19.
The study findings were published in the peer reviewed journal: Frontiers in Molecular Biosciences.
https://www.frontiersin.org/articles/10.3389/fmolb.2023.1270511/full
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
