SARS-CoV-2 Research: Vietnamese Researchers Discover That SARS-CoV-2 Nsp1 Proteins Bind To Ribosome 40S Subunits, Inhibiting mRNA Translation!
SARS-CoV-2 Research - NSP1 Inhibits mRNA Translation Mar 01, 2023 2 years, 11 months, 4 days, 19 hours, 31 minutes ago
SARS-CoV-2 Research: Vietnamese researchers from the Institute for Computational Science and Technology-Vietnam, Duy Tan University-Vietnam and the Polish Academy of Sciences-Poland have in a new study found that SARS-CoV-2 non-structural proteins 1 or Nsp1 proteins bind to ribosome 40S subunits, inhibiting mRNA translation!
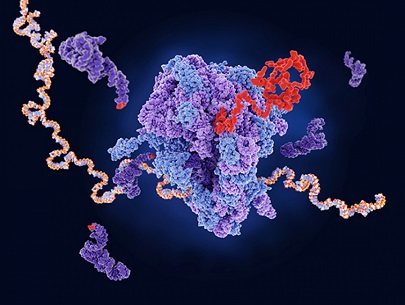
Inhibition or disruption to RNA translation can lead to a reduction in the levels of functional proteins and an increase in deleterious molecules and can affect a cell's normal activities, and/or it may induce checkpoints leading to apoptosis and also substantial effects on gene function by producing misfolded and malfunctioning proteins leading to a variety of medical and health issues!
Past
SARS-Cov-2 Research have already shown that non-structural protein 1 (NSP1) of SARS-CoV-2 is a factor that restricts cellular gene expression and prevents mRNA translation in the ribosome 40S subunit.
https://www.thailandmedical.news/news/breaking-german-study-reveals-that-nsp1-protein-from-sars-cov-2-suppresses-human-host-genes,-long-term-health-implications-expected
However, the molecular mechanism of this phenomenon remains unclear.
In order to clarify this issue, the study team utilized all-atom steered molecular dynamics and coarse-grained alchemical simulations to compare the binding affinity of mRNA to 40S ribosome in the absence and presence of NSP1.
The study findings showed that NSP1 binding to the 40S ribosome dramatically increases the binding affinity of mRNA, which, in agreement with experiment, suggests that NSP1 can stall mRNA translation.
The mRNA translation has been found to be driven by electrostatic mRNA-40S ribosome interactions.
Interestingly, water molecules have been demonstrated to play an important role in stabilizing the mRNA-40S ribosome complex.
The study findings identified the NSP1 residues that are critical in triggering a translation arrest.
The study findings were published on a preprint server and are currently being peer reviewed.
https://www.biorxiv.org/content/10.1101/2023.02.24.529933v1
A past study using all-atom simulation to investigate the interaction between NSP1 and the 40S subunit of the ribosome found that upon NSP1 binding to 40S, the critical switch of Gln158/Glu158 and Glu159/Gln159 residues of SARS-CoV-2/SARS-CoV remodels the interaction pattern between NSP1 and neighboring proteins (uS3 and uS5) and rRNA (h18) lining the exit tunnel. This finding provides a clear picture of how SARS-CoV-2 invades human cells.
https://pubmed.ncbi.nlm.nih.gov/34851631/
&
nbsp;
The study team concluded, “Combining steered molecular dynamics or SMD and alchemical simulations, the association of mRNA with 40S ribosome in the absence and presence of NSP1 was studied. Our SMD results showed that mRNA binds to 40S-NSP1 much more strongly than to 40S ribosome, which is also in the line with the results obtained for the binding free energy using alchemical simulations and the MARTINI model. Our study findings are also in good agreement with the experimental data. The mRNA translation process was found to be driven by the electrostatic interaction between mRNA and 40S ribosome. After entering host cells, NSP1 may bind to the 40S ribosome and inhibit the translation process. Our analysis showed that the NSP1 residues Asp156, Phe157, Gln158, Glu159, Asn160, Trp161, and Asn162 at the interface with mRNA play a key role in triggering translational arrest of the host immune system.”
For the latest
SARS-CoV-2 Research, keep on logging to Thailand Medical News.
