BREAKING! Study Reveals Various ORF10 Protein Mutations In Different Countries And Regions That Specify Different SARS-CoV-2 Pathogenicity Modes!
Source: ORF10-SARS-CoV-2 Dec 08, 2021 4 years, 1 month, 2 weeks, 6 days, 11 hours, 15 minutes ago
To add to all the complications of finding solutions to treat the COVID-19 disease and its long-term health risks, a new study that found not only can COVID-19 ever be classified as a homogenous infection due to the different variants in existence but the fact that there are so many unique protein mutations found on the
ORF10 region of the SARS-CoV-2 that tend to correlate according to geolocations and each uniquely alters the pathogenicity, indicates the only way to treat COVID-19 is via a personalized medicine approach.
The ongoing COVID-19 pandemic caused by the SARS-CoV-2 coronavirus is still wreaking havoc around the world and has made targeting the disease a top priority in medical research and pharmaceutical development.
Genomic surveillance of SARS-CoV-2 mutations is essential for the comprehension of SARS-CoV-2 variant diversity and their impact on virulence and pathogenicity.
The SARS-CoV-2 open reading frame 10 (ORF10) protein interacts with multiple human proteins CUL2, ELOB, ELOC, MAP7D1, PPT1, RBX1, THTPA, TIMM8B, and ZYG11B expressed in lung tissue. Mutations and co-occurring mutations in the emerging SARS-CoV-2 ORF10 variants are expected to impact the severity of the virus and its associated consequences.
The international study team highlighted 128 single mutations and 35 co-occurring mutations in the unique SARS-CoV-2 ORF10 variants. The possible predicted effects of these mutations and co-occurring mutations on the secondary structure of ORF10 variants and host protein interactomes are presented.
The study findings highlight the possible effects of mutations and co-occurring mutations on the emerging 140 ORF10 unique variants from secondary structure and intrinsic protein disorder perspectives.
The study findings were published in the peer reviewed International Journal of Biological Macromolecules.
https://www.sciencedirect.com/science/article/pii/S0141813021025563
It is known that the open reading frame 10 (ORF10) protein of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) interacts with multiple human proteins expressed in lung tissues.
Importantly mutations and co-occurring mutations in the ORF10 strongly impact the virulence and pathogenicity of SARS-CoV-2. The surveillance of ORF10 mutations is therefore crucial for developing potent medical and pharmaceutical interventions against SARS-CoV-2.
The study team examined 140 unique SARS-CoV-2 ORF10 sequences among 202,968 ORF10 sequences retrieved from the National Center for Biotechnology Information (NCBI) database.
The team used the Phobius program to predict the transmembrane topology of SARS-CoV-2 ORF10 protein variants and subjected them to the intrinsic disorder analysis with the PONDR-VSL2 algorithm.
For the study, single mutations in ORF10 proteins were determined using the Virus Pathogen Resource (ViPR), and the frequency distribution of each amino acid in this sequence was determined using standard bioinformatics routine in Matlab.
The study findings revealed unique SARS-CoV-2 ORF10 variants from all six continents including South America, Europe,&a
mp;nbsp;Asia, Oceania, Africa, and North America.
While Africa had the highest percentage of unique variants of ORF10 sequences at 1.27%, the highest frequency of the most unique ORF10 variants was reported in North America.
The study team noticed that all amino acid residues in ORF10 variants from position 1 to 38, except position 18, showed a point-missense mutation. Of a total of 37 residue positions with single mutations, the residue positions 1, 2, 11, 12, 15, 16, 20, 21, 25, 26, 27, 29, 32, 34, and 36 were unique in North America.
Alarmingly however, only single mutations at positions 11 (F11S, F11L) and 16 (L16P) reported in North America appeared detrimental.
It was further found that the unique ORF10 variants from Asia, Africa, and Oceania had several single mutations at common residue positions. However, the two mutations of S23F and S23P in residue S23 were reported across all six continents.
Of significance, the observed unique variations in the SARS-CoV-2 ORF10 variants, especially the co-occurring mutations, appear to be an emerging trend across different continents.
Worryingly these ORF10 variants with co-occurring mutations might be transmitted to other geo-locations in the future when the pandemic-related travel bans end.
Numerous co-occurring mutations in SARS-CoV-2 ORF10 variants were observed in the United States, United Kingdom, India, South Africa, Spain, Germany, Greece, Mexico, and Russia.
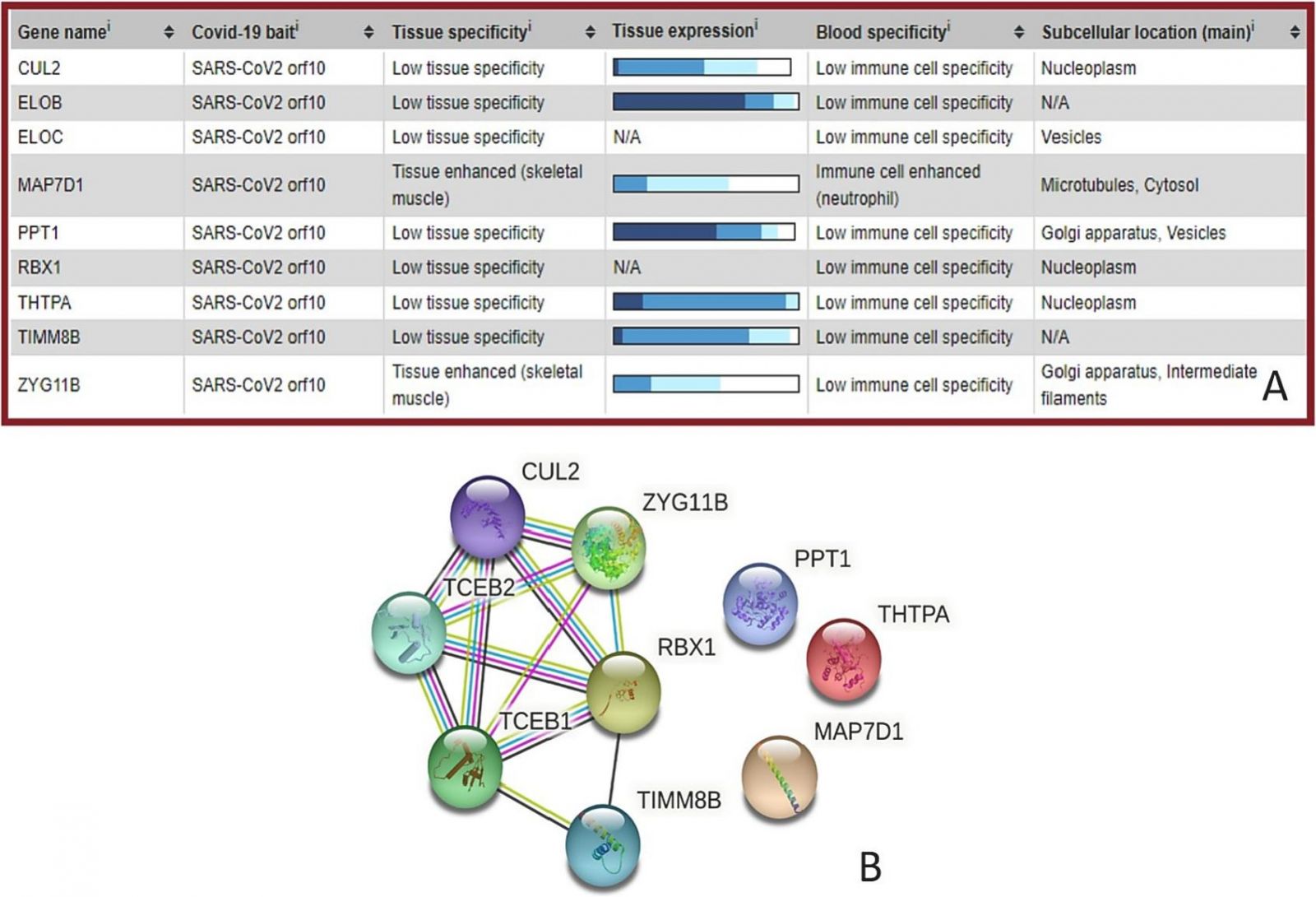 A snapshot summary of the tissue and cellular expression patterns of SARS-CoV-2 ORF10 interacting human proteins, based on transcriptomics and antibody-based proteomics. B: Inter-set protein-protein interaction network of human proteins interacting with ORF10 generated by Search Tool for the Retrieval of Interacting Genes; STRING. STRING produces a network of protein- protein interactions based on predicted and experimentally-validated information on the interaction partners of a protein of interest. In the corresponding network, the nodes correspond to proteins, whereas the edges show predicted or known functional associations. Seven types of evidence are used to build the corresponding network, where they are indicated by the differently colored lines: a green line represents neighborhood evidence; a red line – the presence of fusion evidence; a purple line – experimental evidence; a blue line – co-occurring evidence; a light blue line – database evidence; a yellow line – text mining evidence; and a black line – co-expression evidence.
A snapshot summary of the tissue and cellular expression patterns of SARS-CoV-2 ORF10 interacting human proteins, based on transcriptomics and antibody-based proteomics. B: Inter-set protein-protein interaction network of human proteins interacting with ORF10 generated by Search Tool for the Retrieval of Interacting Genes; STRING. STRING produces a network of protein- protein interactions based on predicted and experimentally-validated information on the interaction partners of a protein of interest. In the corresponding network, the nodes correspond to proteins, whereas the edges show predicted or known functional associations. Seven types of evidence are used to build the corresponding network, where they are indicated by the differently colored lines: a green line represents neighborhood evidence; a red line – the presence of fusion evidence; a purple line – experimental evidence; a blue line – co-occurring evidence; a light blue line – database evidence; a yellow line – text mining evidence; and a black line – co-expression evidence.
Of 22 unique co-occurring mutations observed in different geographical locations, the highest number of co-occurring mutations were reported in March 2021 in a SARS-CoV-2 ORF10 variant from Russia, which exhibited mutations in 14 amino acid residues.
It was also found that the most commonly co-occurring mutations V30L and T38I were first reported in Spain in 2021 and there were no single-point mutations at the residue positions 1 and 2.
However, in the U.S., co-occurring mutations M1K, G2A, Y3D, I4G, N5L, V6Y, F7K, and A8R were reported; however, none of them was a single mutation in the ORF10 variants.
The pathogenic effects of double co-occurring mutations P10S and V30l were surprisingly found to be neutral. Furthermore, V30L, one of the most common mutations in ORF10, was co-occurring with most of the other co-occurring mutations.
Several mutations such as L17I, A8T, V6M, N34P, and F35Q did not appear as a single mutation and only appeared as one of the co-occurring mutations.
Interestingly it was also observed that the effects of co-occurring mutants are qualitatively similar to those of single mutations, as most variability is around a region centered at residue 25 and at N- and C-terminal regions of the protein.
But in co-occurring mutants, as compared to single mutations, scales of changes at the terminal regions are significantly larger. Consequently, these mutations may affect the secondary structure and associated functions of the ORF10 variants.
Alarmingly the study findings show that mutations in the SARS-CoV-2 ORF10 variants, especially co-occurring mutations, are emerging across different continents, with the highest number of single mutations and co-occurring mutations in North America.
The study team says that a possible explanation for this could be that North America is the origin of the largest number of sequenced SARS-CoV-2 isolates. Also, the nations in this continent have implemented different approaches for controlling SARS-CoV-2 transmission locally, thus generating conditions for the locally diversified evolution of SARS-CoV-2.
The research findings also demonstrate that the growth rate of non-synonymous ORF10 mutations is increasingly non-linear, thereby raising questions on the stability or instability of emerging SARS-CoV-2 variants.
Importantly a significant percentage of observed non-synonymous mutations are detrimental, which can alter the expression of the SARS-CoV-2 ORF10 proteins and subsequently affect functional virus-host protein-protein interactions.
Also, it should be noted that co-occurring mutations have significantly changed the secondary structure, particularly the α helix regions of the ORF10 variants.
Most importantly the presence of these mutations can influence the virulence/pathogenicity of SARS-CoV-2 directly or indirectly, which makes continuous surveillance of mutations and their associated effects crucial.
The study team concluded, “The growth rate of emerging non-synonymous mutations in ORF10 proteins is increasing non-linearly, which is certainly alarming with regards to stability or instability of emerging SARS-CoV-2 variants. Due to significant deleterious mutations, expression of the SARS-CoV-2 ORF10 proteins might get altered, a phenomenon, which will affect functional virus-host protein-protein interactions. Also, it was reported that the secondary
structure
α helix regions of the ORF10 variants was changed due to co-occurring mutations. Consequently, virulence/pathogenicity of SARS-CoV-2 may get influenced directly or indirectly.”
Please help to sustain this site and also all our research and community initiatives by making a donation. Your help means a lot and helps saves lives directly and indirectly and we desperately also need financial help now.
https://www.thailandmedical.news/p/sponsorship
For the latest
SARS-CoV-2 Research, keep on logging to Thailand Medical News.

