Researchers Warn That SARS-CoV-2 Envelope Protein Mutations Affects Pathogenicity Of Omicron XBB Sub-lineages
COVID-19 News - SARS-CoV-2 Envelope Protein Mutations – Pathogenicity - Omicron XBB Jul 30, 2023 2 years, 6 months, 2 weeks, 1 day, 54 minutes ago
Study found that T9I mutation was associated with milder variants, while mutations such as P71L, correlated with more severe variants.
COVID-19 News: The emergence of various Omicron variants following the relaxation of COVID-19 restrictions raised concerns about their transmission, immune escape, and pathogenicity. Researchers from the Chinese Academy of Sciences, Shanghai, the University of Chinese Academy of Sciences, Beijing, and Fudan University, Shanghai, investigated the role of envelope protein (2-E) mutations in determining the pathogenicity of Omicron sub-lineages. The study utilized a combination of in vitro and in vivo experiments and analyzed clinical data from different variants. The findings revealed critical insights into the association between 2-E mutations and disease severity, presenting potential pathogenicity markers for SARS-CoV-2.
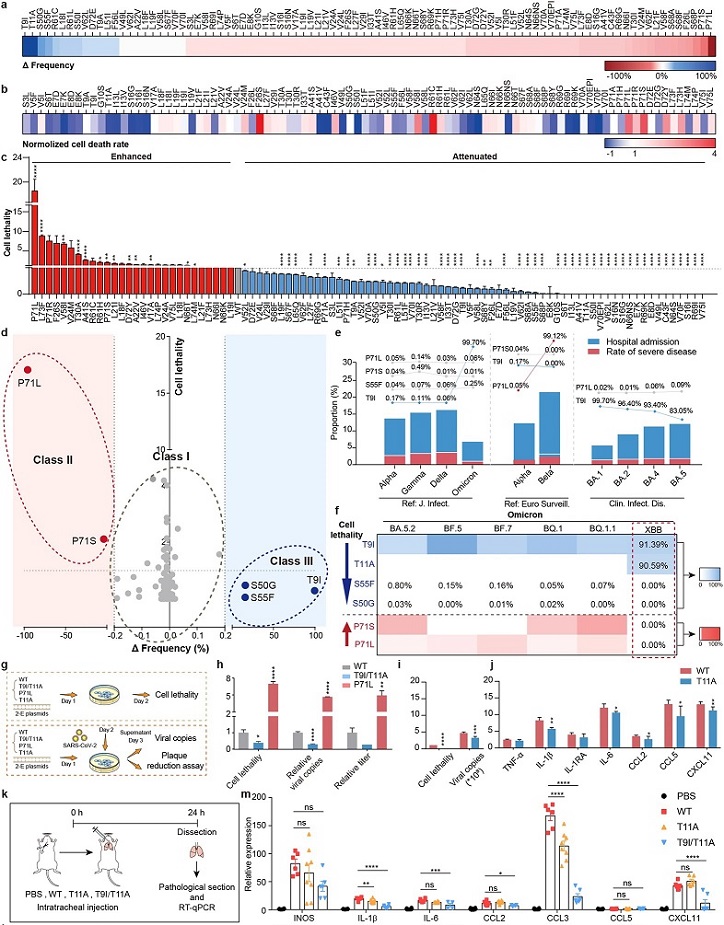 The Δfrequency of 92 2-E mutations. b The normalized cell death rate of 2-E mutations. c The cell lethality of 2-E mutations. d Correlation analysis of Δfrequency and cell lethality. The dotted gray circles represent Class I, the dotted red circles represent Class II, and the dotted blue circles represent Class III (Spearman’s correlation analysis: R2 = 0.33, P < 0.0001). e The quantification of hospitalization rate and disease severity up to Omicron BA.5 and the contribution of Class II and III mutations in tabulate. Pathogenicity of different SARS-CoV-2 variants3,7,8. f Heatmap of 6 key mutation frequencies in Omicron subvariants. g Flow chart of the experiments. h Cell lethality and viral loads for Vero E6 cells after transfection with plasmids as indicated. i, j The activity of 2-E WT and T11A in causing cell lethality, cytokine release, and viral production. k Flow chart of the experiments.
The Δfrequency of 92 2-E mutations. b The normalized cell death rate of 2-E mutations. c The cell lethality of 2-E mutations. d Correlation analysis of Δfrequency and cell lethality. The dotted gray circles represent Class I, the dotted red circles represent Class II, and the dotted blue circles represent Class III (Spearman’s correlation analysis: R2 = 0.33, P < 0.0001). e The quantification of hospitalization rate and disease severity up to Omicron BA.5 and the contribution of Class II and III mutations in tabulate. Pathogenicity of different SARS-CoV-2 variants3,7,8. f Heatmap of 6 key mutation frequencies in Omicron subvariants. g Flow chart of the experiments. h Cell lethality and viral loads for Vero E6 cells after transfection with plasmids as indicated. i, j The activity of 2-E WT and T11A in causing cell lethality, cytokine release, and viral production. k Flow chart of the experiments.
The study team told
COVID-19 News reporters at Thailand Medical News that as Omicron variants continued to spread globally, it became essential to understand the factors influencing their pathogenicity. Previous research had established that mutations in the viral spike (S) protein were associated with enhanced transmission and immune escape. However, the pathogenicity of Omicron variants appeared milder overall, except for the
BA.5 sub-lineage, which showed increased recovery positivity and symptomatic infections. This study aimed to investigate the role of envelope protein (2-E) mutations in determining virus pathogenicity.
Methods
The study team analyzed 92 2-E mutations from five VOCs (Alpha, Beta, Gamma, Delta, and Omicron) based on data from the National Genomics Data Center (NGDC). They calculated the cell lethality of each mutation and correlated it with the frequency of occurrence and clinical severity. Mutations were classified into three groups: Class I (frequency change between -0.20% and 0.20%), Class II (mutations signif
icantly increasing cell lethality), and Class III (mutations with increased frequency but reduced cell lethality). Two mutations, P71L from Class II and T9I from Class III, were selected for further evaluation of inflammatory secretion levels.
Study Findings
Class III mutations, including T9I, were associated with milder variants, while Class II mutations, such as P71L, correlated with more severe variants.
The frequency changes in T9I and P71L appeared to influence virus pathogenicity. Clinical studies on Omicron BA.5 infection confirmed the importance of these mutations in disease severity. Further analysis of newer subvariants, including XBB, revealed the conservation of T9I and a new mutation, T11A. In vitro and in vivo experiments indicated a weakened pathogenicity for the XBB subvariant.
The study presented five potential pathogenicity markers for SARS-CoV-2 based on 2-E mutations. While XBB currently demonstrated reduced pathogenicity, the possibility of future highly pathogenic mutations remains a concern. An evolutionary trade-off between virulence and transmissibility may influence the virus's fitness. Additional research is needed to investigate the role of ubiquitination and degradation in 2-E expression and the relationship between 2-E mutations and the innate antiviral response.
Conclusion
This comprehensive study shed light on the impact of 2-E mutations on the pathogenicity of Omicron sub-lineages. The identified potential pathogenicity markers, including T9I and P71L, provide valuable information for epidemic prevention and preparedness against COVID-19. Further research at the virus level and linking genomic sequencing with clinical patient severity data will strengthen the findings and advance our understanding of the virus's behavior and evolution. As countries continue to face surges in infections, these insights offer a timely warning of potential outbreaks and the need for ongoing vigilance in monitoring viral mutations.
The study findings were published as a correspondence in the peer reviewed journal: Cell Discovery.
https://www.nature.com/articles/s41421-023-00575-7
For the latest
COVID-19 News, keep on logging to Thailand
Medical News.
