COVID-19 News: Study Shows That Newer SARS-CoV-2 Omicron Sub-lineages Are All Evolving To Evade Innate Immunity Via ORF6 Proteins!
Nikhil Prasad Fact checked by:Thailand Medical News Team Jan 17, 2024 2 years, 1 month, 1 week, 3 days, 22 hours, 31 minutes ago
COVID-19 News: The ongoing COVID-19 pandemic has witnessed the emergence of various SARS-CoV-2 variants, with Omicron being the latest and a unique variant of concern (VOC). Recent research conducted by University College London, Imperial College London, and the UKMRC-University of Glasgow Centre for Virus Research sheds light on the evolutionary mechanisms of Omicron subvariants. The study which is covered in this
COVID-19 News report, focuses on the enhanced innate immune suppression exhibited by newer subvariants, specifically BA.4 and BA.5, compared to earlier ones such as BA.1 and BA.2.
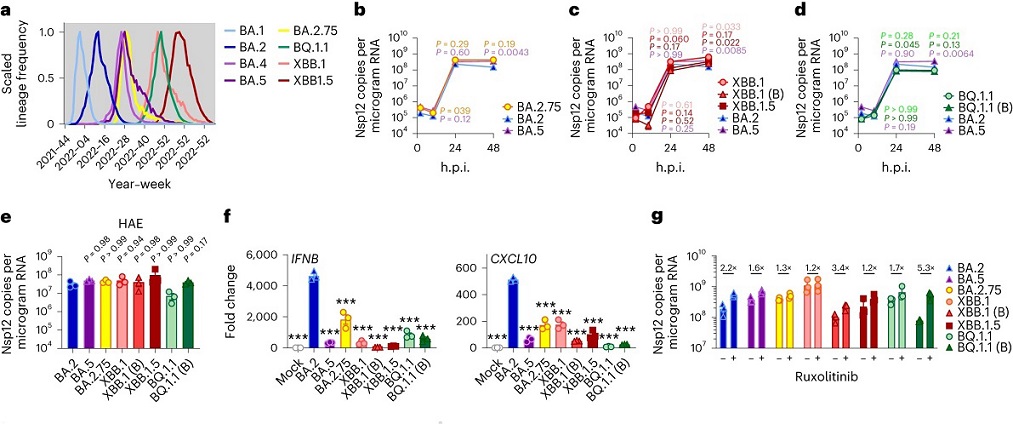 a, Global SARS-CoV-2 variant sequence counts over time (scaled per variant), extracted from CoV-Spectrum using genomic data from GISAID. b–d, Calu-3 cells were infected with 2,000 Nsp12 copies per cell. Replication of Omicron subvariants compared with BA.2 (blue) and BA.5 (purple) measured by Nsp12 copies per microgram RNA is shown for BA.2.75 (yellow; Ο) (b), XBB subvariants (XBB.1: light red, Ο; XBB.1 (B): red, Δ; XBB.1.5: dark red, □) (c) and BQ.1.1 (BQ.1.1: light green, Ο; BQ.1.1 (B): dark green, Δ) (d) isolates. e, HAEs were infected with 1,500 Nsp12 copies per cell and intracellular Nsp12 copies measured at 72 h.p.i. Three biological replicates shown. f, IFNB and CXCL10 expression in Calu-3 cells infected with 2,000 Nsp12 copies per cell of the indicated Omicron subvariants at 24 h.p.i. g, Viral replication of indicated variants in Calu-3 cells in the presence or absence of 5 μM ruxolitinib at 48 h.p.i. Numbers indicate fold change in replication in the presence of 5 μM ruxolitinib.
Evolution of SARS-CoV-2 Variants
a, Global SARS-CoV-2 variant sequence counts over time (scaled per variant), extracted from CoV-Spectrum using genomic data from GISAID. b–d, Calu-3 cells were infected with 2,000 Nsp12 copies per cell. Replication of Omicron subvariants compared with BA.2 (blue) and BA.5 (purple) measured by Nsp12 copies per microgram RNA is shown for BA.2.75 (yellow; Ο) (b), XBB subvariants (XBB.1: light red, Ο; XBB.1 (B): red, Δ; XBB.1.5: dark red, □) (c) and BQ.1.1 (BQ.1.1: light green, Ο; BQ.1.1 (B): dark green, Δ) (d) isolates. e, HAEs were infected with 1,500 Nsp12 copies per cell and intracellular Nsp12 copies measured at 72 h.p.i. Three biological replicates shown. f, IFNB and CXCL10 expression in Calu-3 cells infected with 2,000 Nsp12 copies per cell of the indicated Omicron subvariants at 24 h.p.i. g, Viral replication of indicated variants in Calu-3 cells in the presence or absence of 5 μM ruxolitinib at 48 h.p.i. Numbers indicate fold change in replication in the presence of 5 μM ruxolitinib.
Evolution of SARS-CoV-2 Variants
SARS-CoV-2 has undergone sequential evolution, with variants like Alpha, Delta, and now Omicron dominating globally. Each variant demonstrates unique characteristics that enhance transmissibility, suggesting adaptation to the human host. Previous studies have highlighted the adaptation of Alpha and other VOCs by enhancing the expression of innate immune antagonists like Orf6, N, and Orf9b. However, the emergence of Omicron introduces a shift in selective forces, possibly moving from host adaptation to immune escape from vaccine- and infection-driven memory responses.
Comparative Analysis of Omicron Subvariants
The research compares the replication and host responses of various Omicron subvariants, including BA.1, BA.2, BA.4, BA.5, BA.2.75, and XBB lineages, with the previously dominant Delta variant. The study employs human airway epithelial cells and primary airway cultures to measure viral replication and host responses accurately. Notably, BA.4 and BA.5 demonstrate improved suppression of innate immunity compared to earlier subvariants, hinting at a convergent evolution of enhanced innate immune antagonist expression.
Innate Immune Activation and Viral Replication
The research emphasizes the importance of understanding the phenotypic differences between Omicron subvariants by equalizing the i
nput dose of each variant. The results reveal that BA.4 and BA.5 trigger less innate immune activation than BA.1 and BA.2, even when replication rates are similar. The use of JAK/STAT signaling inhibitors highlights the role of innate immune responses in influencing the infectivity of different subvariants.
Host Response in Primary HAE Cultures
To better mimic the respiratory tract environment, the study extends its analysis to primary human airway epithelial (HAE) cultures. BA.5, despite replicating similarly to BA.2 in HAEs, exhibits reduced innate immune activation. The findings suggest a potential adaptation to reduce innate immune activation as Omicron subvariants evolve.
Temperature Sensitivity and Innate Immune Evasion
The research explores the impact of lower temperatures on Omicron subvariant replication, finding that BA.1 to BA.5 replicate less efficiently at 32°C. Interestingly, despite reduced replication, BA.4 and BA.5 continue to exhibit lower innate immune activation, implying a temperature-independent mechanism of enhanced innate immune evasion.
Role of ORF6 in Enhanced Innate Immune Suppression
The study delves into the molecular mechanisms behind the observed differences in innate immune activation. It identifies increased expression of viral proteins Orf6 and N in BA.4 and BA.5 as a potential explanation. Blocking IFN signaling rescues viral replication, suggesting that Orf6 plays a crucial role in antagonizing innate immune responses.
Consistency Across Omicron Subvariants
As new Omicron subvariants continue to emerge, the research expands its analysis to include BA.2.75, XBB.1, XBB.1.5, and BQ.1.1. The results show a consistent trend of enhanced innate immune evasion, characterized by increased Orf6 expression, across these subvariants. This suggests that evolving innate immune escape mechanisms may be a central feature of Omicron lineage adaptation.
Implications for Understanding Viral Adaptation
The findings contribute to our understanding of how SARS-CoV-2 Omicron subvariants achieve dominance through enhanced innate immune suppression. The study proposes a model where early interactions with host innate immune responses influence viral transmission, with variants that better evade or antagonize innate immunity transmitting more effectively. The research highlights the role of Orf6 in innate immune antagonism and suggests ongoing adaptation towards escaping innate immune mechanisms to enhance transmission success.
Conclusion
The evolving landscape of SARS-CoV-2 variants, particularly the Omicron lineage, continues to pose challenges in understanding the virus's adaptation mechanisms. This comprehensive study provides detailed insights into the enhanced innate immune suppression exhibited by newer Omicron subvariants. The findings underscore the importance of investigating the molecular basis of viral evolution, specifically the role of Orf6 in modulating host responses. As the COVID-19 pandemic unfolds, continuous research efforts are crucial to unravel the intricacies of viral adaptation and inform effective strategies for prevention and treatment.
The study findings were published in the peer reviewed journal: Nature Microbiology.
https://www.nature.com/articles/s41564-023-01588-4
For the latest
COVID-19 News, keep on logging to Thailand Medical News
