Coronavirus News: American Researchers Map Out Which SARS-CoV-2 Mutations Potentially Evade Antibody Recognition. Results Alarming!
Source: Coronavirus News Jan 20, 2021 5 years, 3 weeks, 2 days, 2 hours, 12 minutes ago
Coronavirus News: As the SARS-CoV-2 continues to evolve and mutate to enhance its own survival despite therapeutic interventions such as convalescent plasma, monoclonal and combination monoclonal protocols, vaccines etc, more variants are fast emerging to counter all mankind’s initiatives and its imperative for researchers to identify all possible mutations that appear on the new variants that can potentially evade antibody recognition and neutralizing effects.
.jpg)
In the first 8 months of the COVID-19 pandemic, it must be noted that many stupid, ignorant and non-qualified so called American and British ‘experts’ and ‘virologists’ were making public statements that the SARS-CoV-2 virus was not mutating and even if it did it would most probably lose it potency and simply disappear. We also had lots of stupid journalist and many mainstream and medical American and British media disseminating such misinformation with the aid of American social media platforms and search engines. As a result, there was a lack of attention paid to proper and on time sequencings of all reported cases and also a lack of attention paid to emerging mutations and variants. Thailand Medical news had been issuing warning since the early start of the pandemic about the emergence of more problematic variants but as usual we were often suppressed by the Western world which fortunately is the most badly affected at the moment and hopefully will continue to be so due to their arrogance and nastiness.
By prodding, a team of American researchers from the Fred Hutchinson Cancer Research Center-Seattle, Vanderbilt University Medical Center-Nashville, University of Washington-Seattle and the Howard Hughes Medical Institute-Seattle have in a new study mapped out all the possible mutations in the SARS-CoV-2 coronavirus that can evade antibody recognition.
Antibodies targeting the SARS-CoV-2 spike receptor-binding domain (RBD) are being developed as therapeutics and are a major contributor to neutralizing antibody responses elicited by infection.
Here, the study team describes a deep mutational scanning method to map how all amino-acid mutations in the RBD affect antibody binding and apply this method to 10 human monoclonal antibodies. The escape mutations cluster on several surfaces of the RBD that broadly correspond to structurally defined antibody epitopes.
Interestingly it was found that even antibodies targeting the same surface often have distinct escape mutations. The complete escape maps predict which mutations are selected during viral growth in the presence of single antibodies. They further enable the design of escape-resistant antibody cocktails including cocktails of antibodies that compete for binding to the same RBD surface but have different escape mutations. Therefore, complete escape-mutation maps enable rational design of antibody therapeutics and assessment of the antigenic consequences of viral evolution.
Among the most concerning mutations out of about 4,000 potential mutations, the following are the most concerning: D420, S443, V443, V445, E484, L452, G446, E447,K378, N448, Y449, N450, A474, F486, N487, F490K, S494, A475, G496, Q498, F490, T500, C361, Y365,Y369, N370, F374, T376, N487, K382, S383, P384, F392, K378, R408, A411, K417 and A435.
What i
s alarming is that many of these mutations are becoming more apparent on the newly emerging variants.
The study findings were published on the peer reviewed journal: Cell Host & Microbe.
https://www.cell.com/cell-host-microbe/fulltext/S1931-3128(20)30624-7
At present COVID-19 disease has spread to 191 countries and infected more than 96.2 million individuals. Of these, more than 2.05 million people have lost their lives. The United States remains the nation with the highest number of cases, reaching 24.3 million cases and more than 402,000 American have died from the COVID-19 disease. In the United Kingdom, more than 3.5 million people have been affected and almost 91,500 individuals have died. The infection and death figures as result of COVID-19 is expected to rise exponentially in the coming weeks and even more worse it is expected by the time the third wave arrives in around July or August 2021 where millions could be dying on a daily basis.
Various new strains of the virus have begun to emerge with increased infectivity including the VUI 202012/01 in Britain and 501.V2 in South Africa. This has led to concerns among public health authorities that mutant strains may evade neutralizing antibodies produced by convalescent individuals as well as the present vaccine candidates.
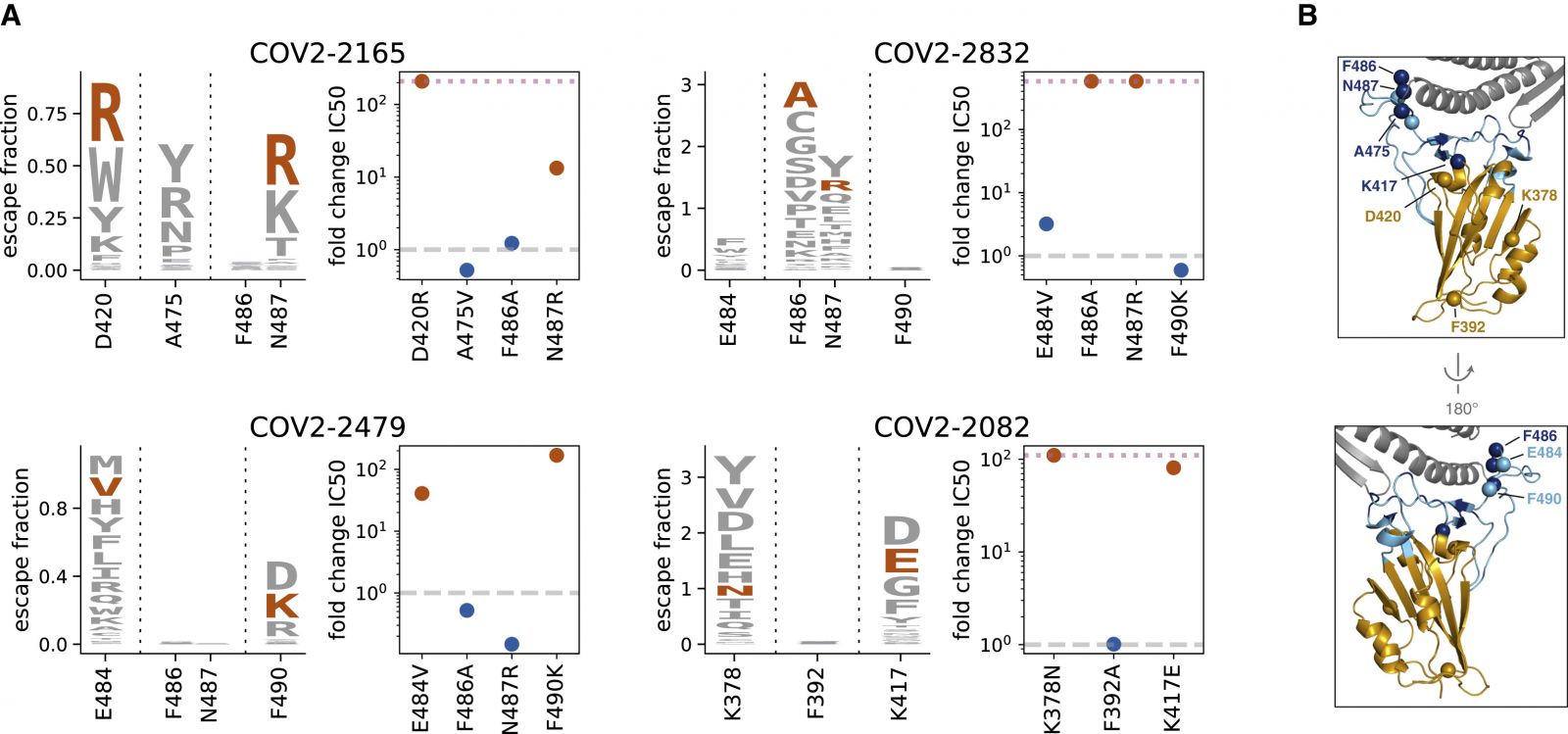 (A) For four antibodies, we validated two mutations that our maps indicated should escape antibody binding and one or two that should not. Logo plots show the escape maps for all tested sites, with the tested mutations expected to escape antibody binding in red. Dot plots show the fold change in neutralization (inhibitory concentration 50%, IC50) relative to wildtype measured using spike-pseudotyped lentiviral particles. Fold changes greater than one (dashed gray line) mean a mutation escapes antibody neutralization. Points in red and blue correspond to mutations expected to mediate or not mediate escape, respectively. Blue letters are not visible in the logo plots due to small escape fractions. Dotted pink lines indicate the upper limit to the dynamic range; points on the line indicate a fold change greater than or equal to this value.Mutations were chosen that had among the largest effects for each of the four antibodies, escaped from multiple antibodies, are present in circulating strains (A475V) or other sarbecoviruses (F490K, E484V) or were surprising in their lack of escape.
(A) For four antibodies, we validated two mutations that our maps indicated should escape antibody binding and one or two that should not. Logo plots show the escape maps for all tested sites, with the tested mutations expected to escape antibody binding in red. Dot plots show the fold change in neutralization (inhibitory concentration 50%, IC50) relative to wildtype measured using spike-pseudotyped lentiviral particles. Fold changes greater than one (dashed gray line) mean a mutation escapes antibody neutralization. Points in red and blue correspond to mutations expected to mediate or not mediate escape, respectively. Blue letters are not visible in the logo plots due to small escape fractions. Dotted pink lines indicate the upper limit to the dynamic range; points on the line indicate a fold change greater than or equal to this value.Mutations were chosen that had among the largest effects for each of the four antibodies, escaped from multiple antibodies, are present in circulating strains (A475V) or other sarbecoviruses (F490K, E484V) or were surprising in their lack of escape.
(B) RBD structure colored with labeled spheres indicating sites where mutation effects on neutralization were validated.
Numerous countries, including Britain and South Africa, are experiencing a successive wave of COVID-19 cases, with skyrocketing numbers being reported in many parts of the world.
In order to better account for these risks, the study team has systematically identified potential SARS-CoV-2 mutations that could evade neutralizing antibody detection.
The SARS-CoV-2 coronavirus acquires mutations that can prevent the antibody response. However, scientists can predict what mutations may emerge as the virus evolves.
The scientist aimed to map how the 4,000 potential mutations to the SARS-CoV-2 spike receptor-binding domain (RBD) can impact antibody binding and neutralization.
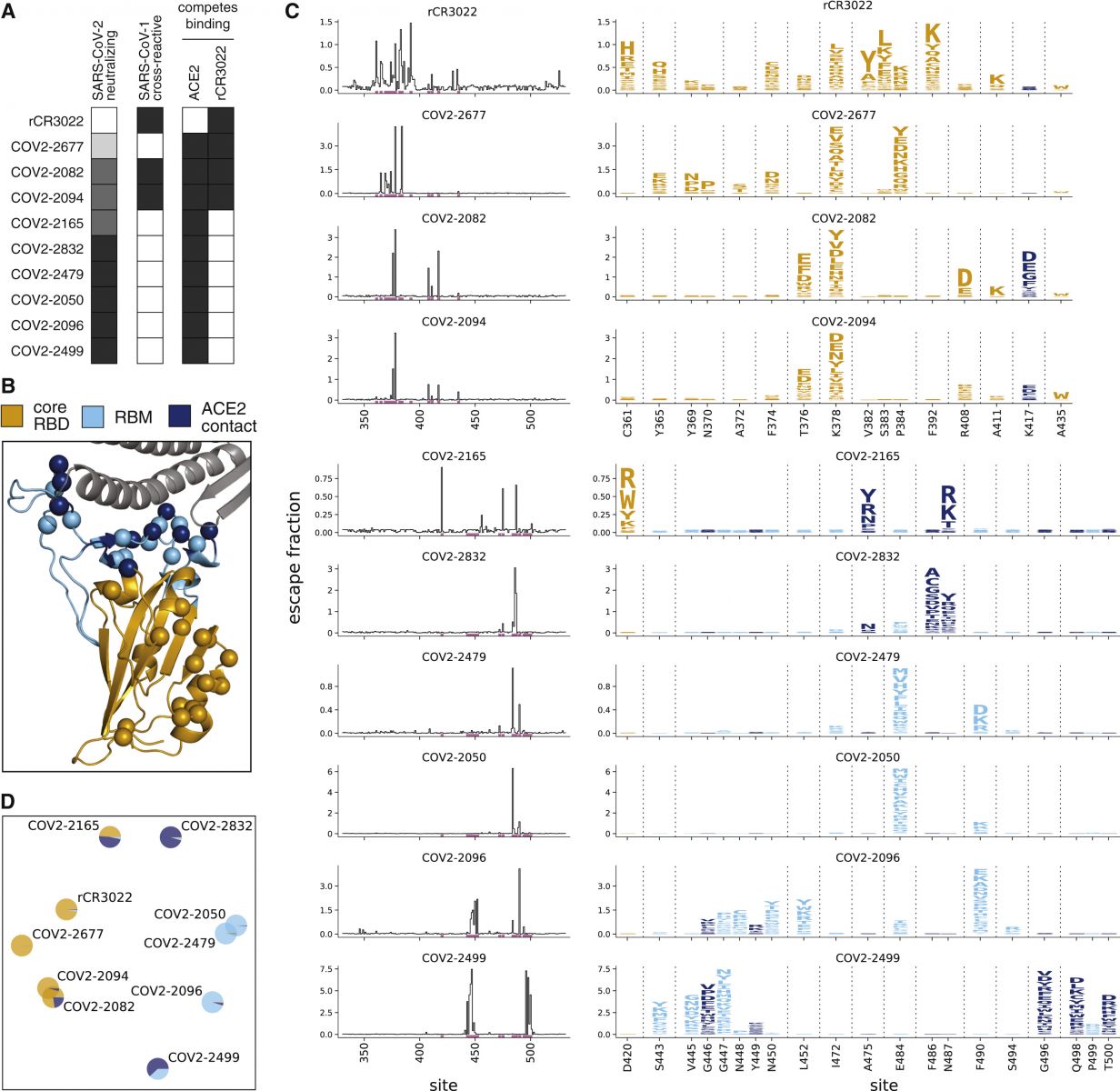 (A) Properties of the antibodies as reported by Zost et al. (2020a). SARS-CoV-2 neutralization potency is represented as a gradient from black (most potent) to white (non-neutralizing). Antibodies that bind SARS-CoV-1 spike or compete with RBD binding to ACE2 or rCR3022 are indicated in black. (B) Structure of the SARS-CoV-2 RBD (PDB: 6M0J; Lan et al., 2020), with residues colored by whether they are in the core RBD distal from ACE2 (orange), in the receptor-binding motif (RBM, light blue), or in direct contact with ACE2 (dark blue). ACE2 is in gray. RBD sites where mutations escape antibodies are indicated with spheres. (C) Maps of escape mutations from each antibody. The line plots show the total escape at each RBD site (sum of escape fractions of all mutations at that site). Sites with strong escape mutations (indicated by purple at bottom of the line plots) are shown in the logo plots. Logo plots are colored by RBD region as in (B). Different sites are shown for the rCR3022-competing antibodies (top four) and all other antibodies (bottom six). For interactive escape maps, see https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_Crowe_antibodies. (D) Multidimensional scaling projection of the escape mutant maps, with antibodies having similar escape mutations drawn close together. Each antibody is shown with chart that uses the color scale in (B) to indicate the RBD regions where it selects escape mutations.
(A) Properties of the antibodies as reported by Zost et al. (2020a). SARS-CoV-2 neutralization potency is represented as a gradient from black (most potent) to white (non-neutralizing). Antibodies that bind SARS-CoV-1 spike or compete with RBD binding to ACE2 or rCR3022 are indicated in black. (B) Structure of the SARS-CoV-2 RBD (PDB: 6M0J; Lan et al., 2020), with residues colored by whether they are in the core RBD distal from ACE2 (orange), in the receptor-binding motif (RBM, light blue), or in direct contact with ACE2 (dark blue). ACE2 is in gray. RBD sites where mutations escape antibodies are indicated with spheres. (C) Maps of escape mutations from each antibody. The line plots show the total escape at each RBD site (sum of escape fractions of all mutations at that site). Sites with strong escape mutations (indicated by purple at bottom of the line plots) are shown in the logo plots. Logo plots are colored by RBD region as in (B). Different sites are shown for the rCR3022-competing antibodies (top four) and all other antibodies (bottom six). For interactive escape maps, see https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_Crowe_antibodies. (D) Multidimensional scaling projection of the escape mutant maps, with antibodies having similar escape mutations drawn close together. Each antibody is shown with chart that uses the color scale in (B) to indicate the RBD regions where it selects escape mutations.
In order to arrive at the study findings, the researchers developed a deep mutational scanning method to map how all amino-acid mutations in the RBD impact antibody binding. They also wanted to apply the method to ten human monoclonal antibodies.
The study team expressed the mutant variants of RBD on the surface of yeast cells and exposed them to ten antibodies, which were extracted from COVID-19-confirmed patients. The team captured the cells that manifested impaired binding to the antibodies and conducted deep sequencing to determine the RBD mutations existing in these cells.
Corresponding author, Dr Jesse D Bloom from the Basic Sciences Division, Fred Hutchinson Cancer Research Center-Seattle and who is also from the Department of Genome Sciences & Medical Scientist Training Program, University of Washington told Thailand Medical news, “The complete escape maps predict which mutations are selected during viral growth in the presence of single antibodies.”
Dr Bloom added, “These maps further enable the design of escape-resistant antibody cocktails-including cocktails of antibodies that compete for binding to the same RBD surface but have different escape mutations.”
The study team also identified some locations on the protein where these escape mutations are grouped. They observed that the mutations tended to differ between various antibodies.
The study team concluded that the complete escape-mutation maps can help develop new therapies and vaccines that can adapt to viral evolution. The researchers also said that they are working on expanding the new approach, to address the largest problem on SARS-CoV-2 evolution.
Dr Allison Greaney, a study co-author added, “The team’s next steps will be to apply this approach to serum from individuals who have been vaccinated against SARS-CoV-2 to see how mutations might reduce binding and neutralization by vaccine-elicited antibodies.”
It should be noted that some viruses, such as measles, are antigenically stable such that immunity from an initial infection or vaccination typically provides life-long protection. Others, such as influenza virus, undergo rapid antigenic drift, such that immunity elicited against one viral strain can be ineffective against that strain’s descendants just a few years later. It remains an open question the extent to which mutations that substantially affect the antigenicity of SARS-CoV-2 will fix during viral evolution. The escape-mutation maps we have generated, as well our methodology for rapidly creating such maps for additional antibodies and sera, should help answer this question by facilitating assessment of the antigenic consequences of mutations observed during viral surveillance.
Please help to support this website and also all our research initiatives by making a small donation. Your help is of tremendous value as it helps saves lives. https://www.thailandmedical.news/p/sponsorship
For the latest
Coronavirus News, keep on logging to Thailand Medical News.
.jpg)

