BREAKING! COVID-19 Test: A New Rapid CRISPR-Based COVID-19 Mobile Phone Diagnostic Test To Debut Soon!
Source: COVID-19 Test Oct 05, 2020 5 years, 4 months, 2 weeks, 6 days, 3 hours, 52 minutes ago
COVID-19 Test: A large multidisciplinary group of researchers from University of California-San Francisco, J. David Gladstone Institutes-San Francisco, University of California-Berkeley, Monash University-Australia and Chan Zuckerberg Biohub-San Francisco have developed an amplification-free CRISPR-based mobile phone assay for direct detection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from nasal swabs.
The assay achieved ~100 copies/µL sensitivity in under 30 minutes and accurately detected a set of positive clinical samples in under 5 minutes.
This new mobile phone based diagnostic tool is not only rapid, portable and accurate but also cheap.
The product is expected to hit the market in not more than a couple of weeks pending U.S.FDA regulatory approvals.
The research findings are published on a preprint server and are pending peer review.
https://www.medrxiv.org/content/10.1101/2020.09.28.20201947v1
The acute challenge of identifying symptomatic, asymptomatic, and presymptomatic carriers of the SARS-CoV-2 coronavirus has been result their delayed isolation and massive global disease spread. Cheap but accurate and convenient COVID-19 testing tools are a critical in controlling the spread of the disease.
At present, the diagnostic gold standard is the quantitative reverse transcription-polymerase chain reaction (RT-qPCR), which is reliable and widely used for screening purposes. However, current data shows that the situation in the United States shows a significant backlog, with 31% of nasal swab-based PCR tests necessitating more than four days to process.
The problem is the same in many countries worldwide if not worse.
This critical factor alone, coupled with a potential quick waning of the immunity after natural infection, highlights the need for rapid, point-of-care testing options that can also reliably detect SARS-CoV-2 genetic material.
Hence, viral diagnostics may also benefit from a successful bacterial strategy to destroy incoming bacteriophages (i.e., viruses that attack bacteria) and build an immunological memory, which is known as CRISPR. The latter can be exploited for the successful and quick detection of SARS-CoV-2.
In order to achieve the high sensitivity needed for testing purposes, current CRISPR diagnostic strategies primarily rely on pre-amplification of target ribonucleic acid (RNA) for subsequent detection by a Cas protein.
In a breakthrough study, a large research group reported the proof of concept of a rapid CRISPR-Cas13a-based diagnostic assay for the direct detection of SARS-CoV-2 RNA.
The research team first needed to optimize Cas13 activation through careful selection of CRISPR RNA complexes, as well as to develop a sensitive and transportable fluorescence detection system for this novel assay.
The easiness of the approach was demonstrated by measuring fluorescence with a mobile phone camera in a close-packed device, comprised of low-cost laser lightning and collection optics.
The study team has also combined complexes of CRISPR RNA and
Cas13 (crRNAs) that target SARS-CoV-2 RNA to enhance the sensitivity and specificity of the proposed diagnostic solutions. They have also directly quantified viral load by utilizing enzyme kinetics.
A major feature of this approach is a successful demonstration of how these combinations of crRNAs can increase the sensitivity of Cas13a direct detection by including more Cas13a (which is a specific CRISPR-Cas13 subtype) per target RNA.
Corresponding author of the study Dr Daniel A. Fletcher from the department of Bioengineering, University of California-Berkeley told Thailand Medical News, "By combining multiple crRNAs to increase Cas13a activation and analyzing the change in fluorescence over time rather than solely endpoint fluorescence, we are able to achieve SARS-CoV-2 viral RNA detection of ~100 copies/µL within 30 minutes."
He further added, "We also correctly identified all SARS-CoV-2 positive patient samples tested within 5 minutes.”
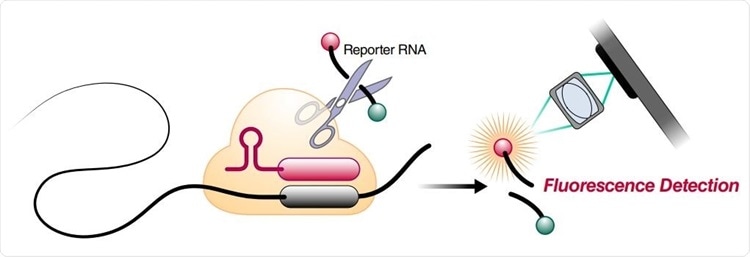 Schematic of a Cas13a (beige)-crRNA (red) RNP complex binding target RNA (black), resulting in activation of the HEPN nuclease (denoted by scissors) domain. Upon target recognition and RNP activation, Cas13a indiscriminately cleaves a quenched-fluorophore RNA reporter, allowing for fluorescence detection as a proxy for Cas13a activation and target RNA. (B) Schematic of the SARS-CoV-2 nucleocapsid (N) gene, and the corresponding locations of each crRNA spacer region.
Schematic of a Cas13a (beige)-crRNA (red) RNP complex binding target RNA (black), resulting in activation of the HEPN nuclease (denoted by scissors) domain. Upon target recognition and RNP activation, Cas13a indiscriminately cleaves a quenched-fluorophore RNA reporter, allowing for fluorescence detection as a proxy for Cas13a activation and target RNA. (B) Schematic of the SARS-CoV-2 nucleocapsid (N) gene, and the corresponding locations of each crRNA spacer region.
Significantly, another key advance is the possibility to translate the fluorescent signal into viral loads directly. Unlike previous diagnostic assays based on CRISPR technology, this one does not require pre-amplification of the viral genome. In other words, by directly detecting the viral RNA (without additional manipulations), we get quantitative RNA measurements instead of a simple, binary positive/negative result.
Importantly, a key facet is that the simple mobile phone-based device can accurately read the direct-detection assay, avoiding the need for a bulky laboratory plate reader. This concept was already tried in previous molecular diagnostic approaches, primarily loop-mediated isothermal amplification.
Dr Fletcher added, "Here we show that direct detection of SARS-CoV-2 RNA with CRISPR-Cas13a and a mobile phone offers a promising option for rapid, point-of-care testing.”
.jpg) Schematic of two different RNPs binding to different locations of the same SARS-CoV-2 RNA, leading to cleavage of the RNA reporter and increased fluorescence.
Schematic of two different RNPs binding to different locations of the same SARS-CoV-2 RNA, leading to cleavage of the RNA reporter and increased fluorescence.
In the last few years, mobile phone cameras have become highly sensitive tools, which (together with GPS, connectivity, and data-processing abilities) have turned them into attractive devices for point-of-care disease diagnosis, especially in low-resource regions.
The study team said, “Combined with mobile phone-based quantification, we can really see the potential in this approach to enable fast, portable, accurate, and low-cost options for point-of-care SARS-CoV-2 screening endeavors.”
In the future direct detection by Cas13a may be quickly modified to target the next emerging respiratory pathogen hopefully in time to help curtail the global spread and another potentially devastating pandemic.
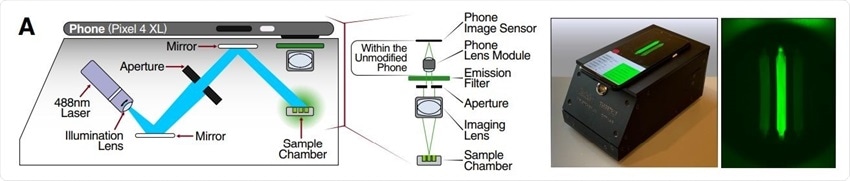 Schematic of mobile phone-based microscope for fluorescence detection showing illumination and image collection components (left). Picture of assembled device used for data collection and sample image taken by the mobile phone camera after running a Cas13a assay (right).
Schematic of mobile phone-based microscope for fluorescence detection showing illumination and image collection components (left). Picture of assembled device used for data collection and sample image taken by the mobile phone camera after running a Cas13a assay (right).
This new mobile phone based direct-detection assay if scaled up, could fulfill the need for a test that provides rapid results and is available to be administered frequently. Other current tests in this category include Abbott Lab’s ID NOW, a PCR-based test, and several antigen tests, such as Quidel’s Sofia 2 SARS Antigen FIA and Abbot Lab’s BinaxNOW Antigen Test.
In the case of influenza, antigen tests span a wide range of sensitivities (e.g., 51–67.5%) (Babin et al., 2011; Chartrand et al., 2012; Chu et al., 2012). Due to the low-to-moderate sensitivity of these tests, the U.S.CDC still recommends re-testing samples that are negative with a more sensitive test, such as RT-qPCR (Green and St George, 2018). Notably, none of the current rapid testing options provides quantitative results, which could help assess vial dynamics and evaluate an individual’s level of infection and progression of disease unlike this new platform.
For more on
COVID-19 Test, keep on logging to Thailand Medical News.


.jpg)
