BREAKING COVID-19 News! 39 Hammerhead-Variant Ribozyme Sequences Identified In SARS-CoV-2!
Nikhil Prasad Fact checked by:Thailand Medical News Team Feb 01, 2024 2 years, 3 weeks, 16 hours, 32 minutes ago
COVID-19 News: As the world grapples with the ongoing challenges posed by the SARS-CoV-2 virus and its variants, a groundbreaking discovery has emerged from Fudan University in Shanghai, China. Researchers have identified 39 Hammerhead-variant ribozyme sequences within the genome of SARS-CoV-2, the virus responsible for the COVID-19 pandemic. This revelation adds a new layer to our understanding of the virus's genomic makeup and its potential implications for viral replication and the overall course of the disease. It is also important note that such a major discovery that has a lot of important implications was only made 4 years after the debut of the SARS-CoV-2 which makes us to realize that there are so many things that we still do not know about this SARS-CoV-2 virus. This
COVID-19 News report touches on the discovery of these Hammerhead-variant ribozyme sequences in SARS-CoV-2.
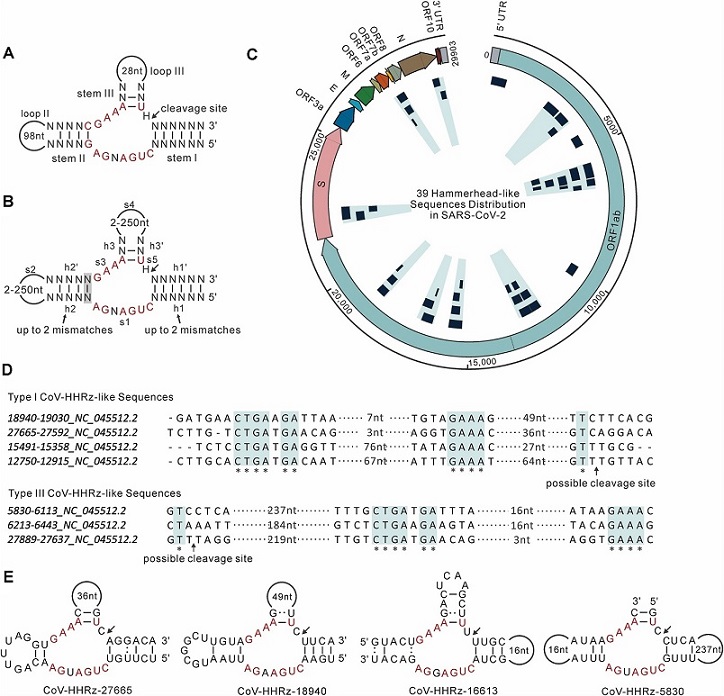 Identification of Hammerhead-variant sequences in the SARS-CoV-2 genome. (A) Covariance model of hammerhead ribozyme. Conserved nucleotides are highlighted in red. The arrowhead indicates the cleavage site. (B) RNABOB search criteria for Type I Hammerhead-variant sequences in this study. The eleven conserved bases are highlighted in red. The grey box shows relaxed base pair adjacent to the catalytic core. Up to two mismatches are allowed in stem I and II. Loop II and III are allowed to be up to 250nt. (C) Distribution of Hammerhead-variant sequences in SARS-CoV-2 genome. The outer circle indicates the SARS-Cov2 genome location (nt), ORFs and 5′ and 3′ UTRs. The light blue radial shadows indicate the cluster of the sequences for the location and coverage of the 39 Hammerhead-variant sequences (as inset navy-blue boxes) in SARS-Cov2 genome. (D) Sequence examples alignment of Hammerhead-variant sequences. The accession numbers and location numbers are shown. Conserved nucleotides are highlighted by blue shading and asterisks. The length of loop II and III are shown. (E) Examples of folded Hammerhead-variant sequences.
Understanding Hammerhead Ribozymes
Identification of Hammerhead-variant sequences in the SARS-CoV-2 genome. (A) Covariance model of hammerhead ribozyme. Conserved nucleotides are highlighted in red. The arrowhead indicates the cleavage site. (B) RNABOB search criteria for Type I Hammerhead-variant sequences in this study. The eleven conserved bases are highlighted in red. The grey box shows relaxed base pair adjacent to the catalytic core. Up to two mismatches are allowed in stem I and II. Loop II and III are allowed to be up to 250nt. (C) Distribution of Hammerhead-variant sequences in SARS-CoV-2 genome. The outer circle indicates the SARS-Cov2 genome location (nt), ORFs and 5′ and 3′ UTRs. The light blue radial shadows indicate the cluster of the sequences for the location and coverage of the 39 Hammerhead-variant sequences (as inset navy-blue boxes) in SARS-Cov2 genome. (D) Sequence examples alignment of Hammerhead-variant sequences. The accession numbers and location numbers are shown. Conserved nucleotides are highlighted by blue shading and asterisks. The length of loop II and III are shown. (E) Examples of folded Hammerhead-variant sequences.
Understanding Hammerhead Ribozymes
Hammerhead ribozymes are small catalytic RNA molecules known for their ability to cleave RNA at specific sites. These ribozymes have been extensively studied for their potential therapeutic applications in manipulating gene expression. Unlike proteins, which are the typical catalytic molecules in biological systems, ribozymes, and particularly hammerhead ribozymes, play a crucial role in various biological functions. They are involved in processes such as RNA splicing, transfer RNA biosynthesis, and viral replication.
The Hammerhead ribozymes consist of a conserved catalytic core. They have the ability to cleave substrate RNA at NUH triplets 3′ to the H. Basically, N represents any nucleotide, U is uracil, and H represents any nucleotide but guanidine, although studies have shown that RNA may be less selective. Typically, they found in the ribosome where they join amino acids together to form protein chains. They can also target specific
host genes!
The SARS-CoV-2 Genome and Its Variants
The SARS-CoV-2 virus, responsible for the COVID-19 pandemic, belongs to the beta coronavirus group within the Coronaviridae family. The virus has undergone various mutations, leading to the emergence of different variants, including the well-known alpha, beta, gamma, delta, lambda, and omicron variants. Despite extensive research on the virus, no ribozymes had been identified in the SARS-CoV-2 genome until now.
Discovery of CoV-HHRz Sequences
The study team identified 39 Hammerhead-variant ribozyme sequences (CoV-HHRz) within the SARS-CoV-2 genome. These sequences exhibit high conservation among SARS-CoV-2 variants but show significant diversity when compared to other coronaviruses. The study delves into the characteristics of these CoV-HHRz sequences, shedding light on their potential role in the viral life cycle.
Bioinformatic Searches and Sequence Characteristics
Bioinformatic searches for Hammerhead ribozyme candidates in the SARS-CoV-2 genome revealed 39 CoV-HHRz sequences. These sequences, categorized into type I, II, and III, are preferentially located in ORF 1ab of the viral genome. The probability of these sequences occurring randomly in the SARS-CoV-2 genome was estimated to be low, emphasizing their potential functional significance.
In Vitro Activity of CoV-HHRz Sequences
To assess the activity of the CoV-HHRz sequences in vitro, the researchers conducted experiments under various conditions. The results showed that these ribozymes exhibited low activity under standard conditions with Mg2+, a common ion for conventional hammerhead ribozymes. However, in the presence of transition metal cations such as Mn2+, Co2+, or Cd2+, cleavage activity was detected. The study highlights the unique ion dependence of these CoV-HHRz sequences, distinguishing them from classical hammerhead ribozymes.
CoV-HHRz Cleavage Sites Coincide with SARS-CoV-2 sgRNA Breakpoints
One intriguing finding is the alignment of CoV-HHRz cleavage sites with the deletion breakpoints observed in subgenomic RNA (sgRNA) transcripts of SARS-CoV-2. This suggests a potential in vivo activity of CoV-HHRz sequences in processing sgRNAs crucial for viral packaging and the virus life cycle. The identification of consistent deletion breakpoints by multiple laboratories using different sequencing strategies and host systems strengthens the evidence for the functional role of CoV-HHRz ribozymes.
Insights from Genome-Wide Chemical Probing
The study utilized genome-wide chemical probing techniques to gain insights into the structural characteristics of SARS-CoV-2 RNA in vitro and in vivo. Chemical probing of the CoV-HHRz-16613 sequence revealed a structured RNA with a hammerhead three-way junction, consistent with its ribozyme nature. However, the study acknowledges challenges in fully characterizing longer CoV-HHRz RNA sequences, hinting at the complexity of their folding patterns.
Conservation Across SARS-CoV-2 Variants and Different Species
An analysis of CoV-HHRz sequence conservation across SARS-CoV-2 variants, including alpha, beta, gamma, delta, lambda, and omicron, demonstrated high sequence identity. Despite the mutation rates observed in other regions of the viral genome, CoV-HHRz sequences maintained remarkable conservation. Comparative analysis with different species, including bat coronavirus (Beta-Cov), pangolin coronavirus, SARS-CoV, MERS-CoV, and PEDV, further highlighted the evolutionary relationships of these sequences.
Conclusion
The discovery of 39 Hammerhead-variant ribozyme sequences in the SARS-CoV-2 genome opens new avenues for understanding the intricacies of viral biology. The unique characteristics of these CoV-HHRz sequences, their ion dependence, and their alignment with sgRNA breakpoints suggest a potential role in the viral life cycle. As research continues, further exploration of these ribozymes may unveil additional layers of complexity in the dynamics of SARS-CoV-2, offering valuable insights for the development of targeted therapeutic strategies and antiviral interventions.
The study findings were published in the peer reviewed journal: Nucleic Acids Research (Oxford Journals).
https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkae037/7595398#438390087
Thailand
Medical News will be doing a follow up article on this study as the implications from this major discovery is simply vast and critical in so many aspects.
For the latest
COVID-19 News, keep on logging to Thailand Medical News.
