BREAKING! California Study Finds That Prior Viral Infections Including That By H3N2 influenza Can Cause COVID-19 Severity Due to Antigenic Interference!
Source: Medical News -Antigenic Interference And COVID-19 Severity Aug 30, 2022 3 years, 5 months, 1 day, 2 hours, 43 minutes ago
During the early part of the COVID-19 pandemic, we were feed with dubious study findings by non-credible researchers that made fake claims that previous influenza infections and even other respiratory infections could offer a certain degree of protection against the SARS-CoV-2 virus and prevent disease severity.
Findings from a new study by medical researchers from the University of California-Irvine-USA not only contradicts these claims but also sounds an alarm that individuals with previous pathogenic infections including that by the H3N2 influenza A virus that prevalent in the United States in 2014, could actually end up with more severe COVId-19 disease as a result of what is known as antigenic interference (AIN).
Antigenic imprinting (AIM) or antigenic interference (AIN) occurs when the immune response adapted for a primary infection instead targets a similar, but not identical, pathogen.
A past study demonstrated the strong association between the presence of antibodies binding to an epitope region from SARS-CoV-2 nucleocapsid, termed Ep9, and COVID-19 disease severity.
https://pubmed.ncbi.nlm.nih.gov/33910993/
Individuals with anti-Ep9 antibodies (Abs) had hallmarks of antigenic interference (AIN), including early IgG upregulation and cytokine-associated injury.
Hence, the immunological memory of a prior infection was hypothesized to drive formation of suboptimal anti-Ep9 Abs in severe COVID-19 infections.
The study team from University of California-Irvine identified a putative primary antigen capable of stimulating production of cross-reactive, anti-Ep9 Abs.
Corresponding author, Dr Gregory A. Weiss, a Professor at the Department of Molecular Biology & Biochemistry, University of California, Irvine told Thailand
Medical News, “Binding assays with patient blood samples directly show cross-reactivity between Abs binding to Ep9 and only one bioinformatics-derived, homologous putative antigen, a sequence derived from the neuraminidase protein of H3N2 influenza A virus. This cross-reactive binding is highly influenza strain specific and sensitive to even single amino acid changes in epitope sequence. The neuraminidase protein is not present in the influenza vaccine, and the anti-Ep9 Abs likely resulted from the widespread influenza infection in 2014.”
The study findings indicate that antigenic interference or AIN from a previous viral infection could underlie some cases of COVID-19 disease severity.
The study findings were published in the peer reviewed journal: PLoS ONE.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0272163
The study if the first to find proof of negative repercussions of immune history due to past pathogenic infections in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.
Importantly, the antibodies adhering to the Ep9 epitope from the SARS-CoV-2 nucleocapsid (N) protein strongly correlate with the severity of the coronavirus disease 2019 (COVID-19) illness.
Typi
cally, when the immune reaction attacks a similar but distinct pathogen rather than the primary infection it was designed for, antigenic interference (AIN) occurs.
Individuals with SARS-CoV-2 but harboring anti-Ep9 antibodies (Abs) exhibited characteristics of AIN, such as early immunoglobulin G (IgG) activation and cytokine-linked injury. Hence, it was likely that the immune memory of a past infection influences inadequate anti-Ep9 Abs development in severe COVID-19 cases.
The study team explored the epitope similarity landscape and cross-reactivity of αEp9 Ab to possibly discover a major antigen triggering an Ab-based immune reaction in αEp9-positive COVID-19 patients. Assays assessed the concentrations of αEp9 IgMs and IgGs in αEp9-positive patients whose plasma was taken at various points post-symptom onset (PSO).
The study tea also used the pBLAST and VAST databases to look for Ep9 structural homologs and sequences. The putative AIN epitope areas were subcloned to phagemids that encoded the sequences as hybrids to the P8 coat protein of the M13 bacteriophage to speed up the adhesion assessments in subsequent assays.
In order to reduce anomaly concentrations and accurately represent the median Ab population among patients, samples from different patients were combined for the preliminary assays.
Cross-reactivity against various potential epitopes was screened utilizing the pooled sample information.
Subsequently, Ep9 homolog binding to αEp9 Abs was examined using phage enzyme-linked immunosorbent assays (ELISAs).
The study team next studied the selectivity of αEp9 Abs adhering to neuraminidase (NA) from various viral strains.
The study team further examined if EpNeu from the H3N2 influenza A sequence and Ep9 epitopes attach to the same Abs.
Shockingly, the study found that αEp9 IgG titers of a patient appeared to increase as early as one day PSO, which was commensurate with the characteristics of AIN following a previous infection. Comparable IgG concentrations were seen in the patient group for more than four weeks; as a result, αEp9 IgG titers spiked and stayed high. Likewise, αEp9 IgM levels among patients at different PSO periods were comparable. Compared to similar Ep9 IgG levels, the indications for Ep9 IgM titers were much lower; this discrepancy may be due to reduced IgM quantity, affinity, or both.
Just as critical, the finding that both IgM and IgG antibodies target the Ep9 epitope implies that numerous antibodies with comparable binding characteristics may exist across COVID-19 patients.
Hence the study team pointed out the anti-Ep9 paratopes as a part of an Abs population in sera.
Also, SARS-CoV-2 Ep9 and a corresponding SARS-CoV-1 epitope having 90% similarity bind solely to plasma from Ep9-positive COVID-19 patients, corroborating earlier reported results.
The study team said that due to the limited distribution of SARS-CoV-1 in the United States (US), the αEp9 Ab preference for SARS-CoV-1 was unlikely to induce SARS-CoV-2 AIN.
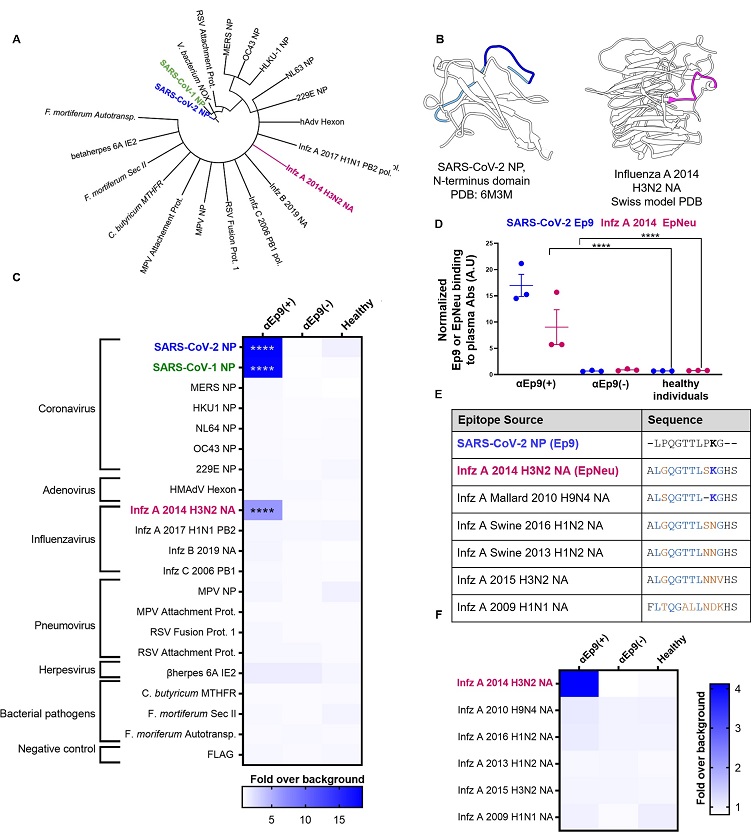 Potential OAS epitopes for binding αEp9 Abs suggested by bioinformatics and tested by phage ELISA.
Potential OAS epitopes for binding αEp9 Abs suggested by bioinformatics and tested by phage ELISA.
(A) Cladogram depicting sequence homology of the Ep9 sequence from SARS-CoV-2 to the bioinformatics-identified, closest homologs. Sequence alignments used pBLAST and VAST, and the cladogram was generated by iTOL [13]. (B) Structures of SARS-CoV-2 NP RNA binding domain (PDB: 6M3M) and the influenza virus (Infz) A 2014 H3N2 NA protein (modeled by SWISS-Model [14]). SARS-CoV-2 NP highlights Ep9 residues (light and dark blue) and the region homologous region to EpNeu (dark blue). The depicted model of Infz A 2014 H3N2 NA highlights the EpNeu putative antigen (pink). (C) ELISAs examined binding of phage-displayed potential OAS epitopes to total Ig from three sets of pooled plasma from five αEp9(+) patients, or five αEp9(−) patients. Pooled plasma from healthy individuals was an additional negative control. The colors of the heat map represent the mean binding signal normalized to phage background negative controls (signal from phage without a displayed peptide). (D) Expansion of data from panel C shows ELISA signals from the independently assayed individual pools shows results from the individual pools (****p <0.0001 for a two-way ANOVA comparing binding of phage-displayed epitopes listed in panel C to different groups of pooled plasma, ad hoc Tukey test). (E) Amino acid sequence alignment of the closely related Infz A NA homologs of EpNeu from pBLAST [11]. Blue and orange residues represent conserved and mismatched amino acids, respectively, relative to Ep9. Bolded residues are important for epitope recognition by αEp9 Abs. (F) Using EpNeu as the search template to generate homologous sequences (shown in panel E), ELISAs examined EpNeu homologs’ binding to pooled plasma from αEp9(+), αEp9(−), or healthy individuals. The data are represented as described in panel C (****p <0.0001 for two-way ANOVA c phage-displayed epitopes, ad hoc Tukey and Dunnett’s test as shown).
However, a putative epitope from the H3N2 influenza A strain NA protein circulated during 2014, named EpNeu by the study team, was identified from the possible epitope panel.
Interestingly, EpNeu was bound by plasma from three separate pools of αEp9-positive SARS-CoV-2 patients but not by plasma from αEp9-negative subjects or healthy people. Despite the 38% amino acid sequence homology between EpNeu and Ep9, other potential epitope sites with noticeably higher homology did not bind to αEp9-positive plasma.
It was also noted that no EpNeu homologs were linked to Abs from αEp9-positive patients, despite having just one residue variation or a 92.3% similarity to EpNeu.
However, one EpNeu amino acid change, K142N in an H1N2 swine flu strain from 2016, significantly reduced binding preference to Abs from patients who were αEp9-positive. A 2010 H9N4 avian influenza A virus epitope that lacked S141 residue but still had conserved K142 significantly decreased adherence to Abs from αEp9-positive patients. Thus, the team suggested that S141 and K142 were essential for αEp9 Ab binding.
The study team stated that αEp9 Abs also bind to the EpNeu epitope. They also predicted that the mean number of Abs in each pool, namely αEp9 and EpNeu, would be comparable, negating the need for further optimization.
The study team also discovered a potential primary antigen that could promote the generation of cross-reactive, anti-Ep9 Abs. Direct cross-reactivity among Abs adhering to Ep9 and just one bioinformatics-derived similar presumptive antigen, a sequence generated from the H3N2 influenza A virus NA protein, was demonstrated by binding experiments utilizing patient blood samples.
It should also be noted that this cross-reactive attachment was very strain-specific for the influenza virus and susceptible to even single amino acid alterations in the epitope sequence.
The study team mentioned that the influenza vaccine did not harbor the NA protein, and the extensive influenza infection during 2014 perhaps resulted in anti-Ep9 Abs.
The study team concluded that some cases of COVID-19 disease severity linked to αEp9 Abs might be caused by AIN from a prior H3N2 influenza A virus infection.
Although several factors may contribute to illness severity during COVID-19, the study findings imply that a subgroup of COVID-19 patients' dependence on elevated titers of imprinted influenza Abs could indicate a less functional immune reaction and, as a result, more severe illness outcomes.
The study team added that future studies should look at the relationship between the prevalence of the H3N2 2014 influenza virus and severe SARS-CoV-2 infection in the United States.
Furthermore, the study team noted that this association could be analyzed via health networks recording influenza infections.
The study team added that the prediction of AIN-based immune reactions and illness outcomes in upcoming infections may be possible by examining Ab cross-reactivity and epitope conservation. Also, recognizing benign, beneficial, or detrimental AIN routes could also play a role in a more effective vaccine design.
It should also be noted that humoral immunity to hCoVs (NL63 and 229E, respiratory syncytial virus, cytomegalovirus and herpes simplex virus-1, has also been associated with more severe COVID-19 disease.
https://www.mdpi.com/2075-1729/11/4/298
https://pubmed.ncbi.nlm.nih.gov/32994364/
https://pubmed.ncbi.nlm.nih.gov/32908997/
It would also be interesting to see if antigenic interference also exists and contributes to disease severity when individuals who had been previously infected with the previous SARS-CoV-2 variants get re-infected with the newer emerging variants or in the case of those who received the COVID-19 jabs with the spike proteins of the original wildtype Wuhan strain.
For more on
Antigenic Interference And COVID-19 Severity, keep on logging to Thailand
Medical News.

